Transforming growth factor beta receptor II polymorphisms are associated with Kawasaki disease
Article information
Abstract
Purpose
Transforming growth factor beta receptor 2 (TGFBR2) is a tumor suppressor gene that plays a role in the differentiation of striated cells and remodeling of coronary arteries. Single nucleotide polymorphisms (SNPs) of this gene are associated with Marfan syndrome and sudden death in patients with coronary artery disease. Cardiovascular remodeling and T cell activation of TGFBR2 gene suggest that the TGFBR2 gene SNPs are related to the pathogenesis of Kawasaki disease (KD) and coronary artery lesion (CAL).
Methods
The subjects were 105 patients with KD and 500 healthy adults as controls. Mean age of KD group was 32 months age and 26.6% of those had CAL. We selected TGFBR2 gene SNPs from serum and performed direct sequencing.
Results
The sequences of the eleven SNPs in the TGFBR2 gene were compared between the KD group and controls. Three SNPs (rs1495592, rs6550004, rs795430) were associated with development of KD (P=0.019, P=0.026, P=0.016, respectively). One SNP (rs1495592) was associated with CAL in KD group (P=0.022).
Conclusion
Eleven SNPs in TGFBR2 gene were identified at that time the genome wide association. But, with the change of the data base, only six SNPs remained associated with the TGFBR2 gene. One of the six SNPs (rs6550004) was associated with development of KD. One SNP associated with CAL (rs1495592) was disassociated from the TGFBR2 gene. The other five SNPs were not functionally identified, but these SNPs are notable because the data base is changing. Further studies involving larger group of patients with KD are needed.
Introduction
Kawasaki disease (KD) is one of the most common vasculitis in childhood1). The etiology of KD remains unknown. Many studies suggest that KD is caused by immunologic response or an agent that can be transmitted. Also, genetic factors appear to contribute to the pathogenesis of KD, as evidenced by the increased frequency of the disease in Asian and Asian-American populations, and among family members of an index case1-4).
In the pathogenesis of coronary arterial inflammation in KD, T cell activation and regulation play important roles5-7).
Transforming growth factor beta (TGF-β) receptor 2 (TGFBR2) gene is a putative tumor suppressor gene that has been implicated in several malignancies8). The gene keeps cells from growing and dividing too fast or in an uncontrolled way. It plays a role in the formation of the extracellular matrix. It also has a role in the differentiation of striated muscle cells and in the remodeling of the coronary artery. In the cardiovascular system, TGF-β can induce neoangiogenesis, cardiomyocyte hypertrophy, calcification, and fibrosis9,10). Mutations in this gene can cause a thoracic aortic aneurysm and dissection11) and Loeys-Dietz aortic aneurysm syndrome12). Single nucleotide polymorphisms (SNPs) of TGFBR2 are associated with Marfan syndrome13), abdominal aortic aneurysm14) and sudden death in patients with coronary artery disease15).
This cardiovascular remodeling as well as T cell activation of the TGF-β gene suggest that the TGF-β gene may be related to the pathogenesis and coronary artery lesion (CAL) of KD. In a recent study, genetic variations in the TGF-β pathway were reported to influence KD susceptibility, disease outcome, and response to therapy16)
The present study was undertaken to address the hypothesis that the TGFBR2 gene is related to the pathogenesis of Kawasaki disease and CAL.
Materials and methods
1. Subjects
KD patients were selected from the Department of Pediatrics, Kyung Hee University Hospital at Gangdong and Kyung Hee University Medical Center from June, 2003 to December, 2005, who were diagnosed as KD according to the guidelines of the Japanese Kawasaki Disease Research Committee17).
CAL were defined when either the right or the left coronary arteries had a diameter of ≥3 mm in children younger than 5 years or ≥4 mm in children older than 5 years, or a diameter >1.5 times that of an adjacent vessel18).
The subjects included 105 patients with KD and 500 controls. The controls were healthy adults who did not have a history of KD. Mean age of the control group was 41 years old (median age, 39.4 years old). The patients are 34 girls and 71 boys. Mean age of patients was 32 months (median age, 25 months) and 26.6% of them had coronary artery lesions.
2. SNP selection and genotyping
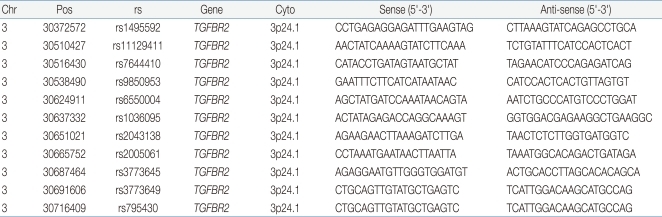
We selected eleven SNPs: rs 1495592, rs11129411, rs7644410, rs9850953, rs6550004, rs1036095, rs2043138, rs2005061, rs3773645, rs3773649, and rs795430 within the TGFBR2 gene region using Illumina Sentrix Array Matrix chip and Helix Tree. The sequences of the forward and reverse primers are summarized in Table 1.
Polymerase chain reaction (PCR) was performed in 20 µL volumes with 10× buffer, 2.5 mM dNTP, and 10 pmol of the forward and reverse primers for rs1495592, rs11129411, rs7644410, rs9850953, rs6550004, rs1036095, rs2043138, rs2005061, rs3773645, rs3773649, and rs795430 and 1 U of Taq DNA polymerase. DNA PCR was performed by 40 cycles of denaturation at 94℃ for 30 seconds, annealing at 58℃ for 30 seconds, and extension at 72℃ for 30 seconds.
3. Statistical analysis
The chi-square (χ2) test was used to determine the Hardy-Weinberg equilibrium between each genotype and each individual by SNPstats. If needed, the Fisher's exact test was used.
We compared between control group and KD patient group to determine whether the SNPs of the TGFBR2 gene were associated with the development of KD. Also, we compared the normal coronary artery group and CAL group to determine whether the SNPs of the TGFBR2 gene were associated with CAL in KD.
Multiple logistic regression models were calculated for odds ratio (OR), 95% confidence interval (CI), and corresponding P values. Statistical significance was set at a P<0.05.
Results
The genetic association study between eleven SNPs of the TGFBR2 gene and susceptibility to KD was investigated. As shown in Table 2, genotype frequencies of eleven SNPs in TGFBR2 gene showed in the KD group and the control group. We analyzed the association between TGFBR2 polymorphisms and susceptibility to KD by the logistic regression model. Logistic regression analysis was revealed that three SNPs of the TGFBR2 gene was associated with KD (rs1495592, OR=1.48, 95% CI=1.07 to 2.05, P=0.019 in log-additive model [C/C vs. T/C vs. T/T], OR=2.12, 95% CI=1.30 to 3.46, P=0.0019 in dominant model [C/C vs. T/C and T/T]; rs6550004, OR=0.56, 95% CI=0.32 to 0.96, P=0.026 in log-additive model [A/A vs. A/C vs. C/C]; rs795430, OR=0.39, 95% CI=0.17 to 0.90, P=0.016 in recessive model [C/C and T/C vs. T/T]).

Genotype Frequencies of Polymorphisms of TGFBR2 Genes in Patients with Kawasaki Disease and Control Subjects
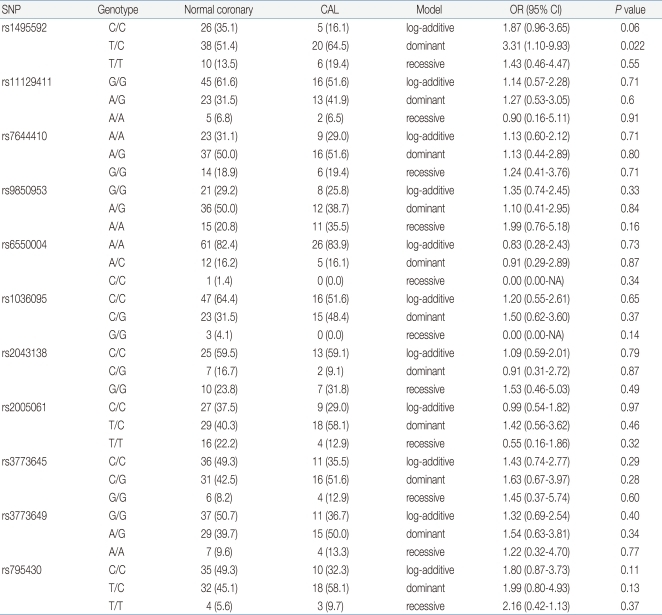
We also compared KD with normal coronary artery group and KD with CAL group. As shown in Table 3, one SNP was associated with CAL in KD (rs1495592, OR=3.31, 95% CI=1.10 to 9.93, P=0.022 in dominant model [C/C vs. T/C and T/T]).
Linkage disequilibrium (LD) block between rs3773645 and rs3773649 was determined using Haploview ver. 4.2 (Fig. 1). Since the LD block was made (D'=0.99), the analysis of haplotypes was performed. However, the association was not observed (P>0.05, data not shown).
Discussion
Various gene polymorphisms are associated with pathogenesis of KD. They involve the inositol 1,4,5-triphosphate 3-kinase C gene5), HLA genes (B5, B44, Bw51 etc.)19,20), angiotensin-1 converting enzyme gene 21) and genetic variation in the chemokine receptor CCR5 and its major ligand CCL3L122). Genes that are associated with CAL in KD are the mannose-binding lectine gene23), promoter of the CD 14 gene24), macrophage migration inhibitory factor-173 polymorphism25), vascular endothelial growth factor (VEGF) and VEGF receptor gene26, 27).
TGFBR2 is a member of the Ser/Thr protein kinase family and the TGF-β receptor subfamily28). This gene is located on chromosome 3 (3p22). It has a polyadenine mononucleotide repeat in its coding region29). The encoded protein is a transmembrane protein that has a protein kinase domain, forms a heterodimeric complex with another receptor protein, and binds to TGF-β. This receptor/ligand complex phosphorylates proteins, which then enter the nucleus and regulate the transcription of a subset of genes related to cell proliferation.
TGFBR2 is a putative tumor suppressor gene and plays a role in the differentiation of striated cells and remodeling of coronary arteries. Mutation of this gene is associated with Marfan syndrome, Loeys-Dietz syndrome, familial thoracic aortic aneurysms and dissections and sudden cardiac arrests in patients with coronary artery disease13-15). KD is known to be associated with lesion of small-to-medium size arteries in most of the patients. But thoracic aortic aneurysms as the complication of KD are reported in 0.9% of patients with KD30) and aortic root dilatations are reported in 8% of patients with KD31). Also, there is another report that a 33 years-old man suffered from incomplete KD had a coronary artery bypass surgery because of the multiple-vessel coronary artery disease. He had an abdominal aortic replacement due to an abdominal aortic aneurysm after 14 years32). We also have experienced the patient with KD complicated with abdominal aortic aneurysm in infant. So, we suggest that we should consider the involvement of large systemic artery in KD although it rarely occur.
TGF-β signaling is critical for the differentiation of smooth muscles into quiescent cells expressing a full repertoire of contractile proteins. Heterozygous mutations in TGFBR2 disrupt TGF-β signaling that makes genetic conditions that predispose to thoracic aortic aneurysm and dissections. TGF-β signaling through the TGFBR2 receptor in endothelial cells plays an important role in cardiac development. It promotes myocardial fibrosis and remodeling with coronary artery disease33).
TGF-β is a protein that controls proliferation, cellular differentiation, and other functions in most cells. It may also act as an antiproliferative factor through DAXX (a death-domain-associated protein) pathway and SMAD (homologs of both the drosophila protein, mothers against decapentaplegic [MAD] and the Caenorhabditis elegans protein SMA) pathway. In the DAXX pathway, TGF-β may trigger apoptosis via the death associated protein 6 (DAXX adapter protein). In the SMAD pathway, activated TGF-β binds to TGFBR2. TGFBR2 recruits and activates the type 1 receptor. The activated type 1 receptor phosphorylates SMAD molecules. These activated SMADs form a complex with Smad4. They enter the nucleus and regulate gene transcription.
Therefore, mutation of TGFBR2 gene can cause an alteration of TGF-β signaling, which may be implicated in the pathogenesis of KD. Genetic variation in the TGF-β pathway (TGFB2, TGFBR2, and SMAD3) may influence KD susceptibility, disease outcome, and response to therapy16). The serum levels of TGF-beta 1 are decreased in patients with KD34).
In this study, we compared eleven SNPs in TGFBR2 between a KD group and control group or KD with normal coronary artery group and KD with CAL group. Three SNPs (rs1495592, rs6550004, rs795430) were associated with the development of KD. One SNP (rs1495592) was associated with CAL in patients with KD.
But, as the data base had been changed, only six of these eleven SNPs have remained as SNPs of the TGFBR2 gene (rs6550004, rs1036095, rs2043138, rs2005061, rs3773645, and rs3773649). One SNP (rs6550004) of three SNPs (rs1495592, rs6550004, rs795430) that were associated with development of KD remained as SNPs of the TGFBR2 gene. The function of the other five SNPs is not known, but these SNPs are notable because the data base is changing. Their function should be the subject of further study.
The limitation of this study was that the size of the study population was relatively small. Further large-scale studies are required to confirm the relationship between mutation of the TGFBR2 gene and KD and its complication, CAL. We studied 11 SNPs of the TGFBR2 gene. Studies involving more SNPs of TGFBR2 gene would be anticipated to discover more SNPs associated with KD.
In conclusion, we have shown that the SNP rs6550004 of the TGFBR2 gene may lead to susceptibility to KD. One SNP (rs1495592) was associated with CAL in KD patients, but this SNP has since been dissociated from the TGFBR2 gene.