Big data analysis and artificial intelligence in epilepsy – common data model analysis and machine learning-based seizure detection and forecasting
Article information
Abstract
There has been significant interest in big data analysis and artificial intelligence (AI) in medicine. Ever-increasing medical data and advanced computing power have enabled the number of big data analyses and AI studies to increase rapidly. Here we briefly introduce epilepsy, big data, and AI and review big data analysis using a common data model. Studies in which AI has been actively applied, such as those of electroencephalography epileptiform discharge detection, seizure detection, and forecasting, will be reviewed. We will also provide practical suggestions for pediatricians to understand and interpret big data analysis and AI research and work together with technical expertise.
Key message
· Big data analysis, such as common data model and artificial intelligence, can solve relevant questions and improve clinical care.
· Recent deep learning studies achieved 0.887–0.996 areas under the receiver operating characteristic curve for automated interictal epileptiform discharge detection.
· Recent deep learning studies achieved 62.3%–99.0% accuracy for interictal-ictal classification in seizure detection and 75.0%–87.8% sensitivity with a 0.06–0.21/hr false positive rate in seizure forecasting.
Graphical abstract.
Introduction
Digital transformation of medical information has changed the hospital environment tremendously over the last 20–30 years. With the implementation of the order communication system and laboratory information system (LIS), medical order slips and lab result papers started to disappear from the hospital environment. In addition, the digitalization of image results and continuous physiologic monitoring data such as electroencephalograms (EEGs) made imaging films and EEG or electrocardiogram strips obsolete. This change, called a “paperless hospital,” was not just a transformation of data storage; rather, it has ushered in a new era of digitalizing all different kinds of patient care data. Most of the tertiary hospitals in the Republic of Korea utilize some form of hospital information system, and there are even cloud systems that local clinics can use without constructing expensive and labor-intensive computer servers.
Advances in computer systems, data storage, and computing superpowers have made big data analysis and artificial intelligence (AI) applications possible in various parts of science. AI is utilized in many regions in medicine, such as diagnosis, treatment decision support, prognosis forecasting, and even public health decision-making [1-3]. Rapidly expanding research using big data analysis and AI applications can provide new opportunities and solutions to unsolved relevant clinical and basic research questions. A basic understanding of big data analysis and AI will help pediatricians follow fast changes in clinical and research fields that are already underway. This review will briefly introduce big data analysis and AI and detail their strengths and limitations. Next it will introduce a common data model (CDM) and CDM applications in clinical research. After that we will review the most recent studies in the field of epilepsy that have applied deep learning (DL); AI in EEG for spike detection, seizure detection, and seizure forecasting. Finally, we will make suggestions and describe relevant issues for pediatricians for future studies.
Epilepsy
Epilepsy is a condition characterized by a predisposition to recurrent spontaneous seizures. There are various kinds of epileptic seizures (focal, generalized, and unknown) and various epilepsies and epilepsy syndromes [4]. Its causes were recently classified as structural, genetic, infectious, metabolic, immune, and unknown [5]. It is well known that epilepsy is a heterogeneous and diverse condition. In fact, it is among the most common chronic neurological conditions in the field of pediatric neurology and affects an estimated 50 million people worldwide [6,7].
Seizure control using various antiseizure medications (ASMs) without adverse reactions is the initial goal of epilepsy management. More than 50% of patients achieve and maintain seizure freedom with ASM treatment, while the remaining 30%–40% have medication-refractory epilepsy [8]. Various comorbidities, such as behavioral and psychiatric issues, are of significant concern in epilepsy. There are relevant issues such as learning and cognitive development in the pediatric population and specific subjects for women with epilepsy and elderly patients. Due to its chronic nature, large amounts of medical records, prescription data, and laboratory results, such as brain imaging and electrophysiological studies, are produced. These data are used as substrates for big data analysis and AI algorithm construction. Some examples of the desired goals of such studies include improving seizure control, decreasing ASM adverse events, and predicting outcomes. Another area of active research is medication-refractory seizures. Uncontrolled seizures are detrimental to both patients and their caregivers, as seizures are unpredictable events that cause significant physical and mental harm. Significant efforts have been made to identify the causative lesions in the brain to improve surgical outcomes and detect and forecast seizures to prevent further seizures or subsequent injury. Recent advances in machine learning (ML) have helped improve outcomes in handling big data, such as continuous EEG monitoring data using scalp and intracranial EEG [9].
Big data analysis using CDM
1. Big data analysis
In 1997, Michael Cox and David Ellsworth first used big data to describe large volumes of scientific data for visualization [10]. According to Gartner, big data is traditionally defined as 3V (volume, velocity, and variety). This means high-volume, high-velocity, and high-variety information assets that demand cost-effective innovative forms of information processing that enable enhanced insight, decision-making, and process automation. With the advent of big data in academic and industrial research, big data terminology is continuously changing [11]. IBM divides big data into 4 dimensions, adding veracity elements to Gartner’s 3V, known as 4V (volume, velocity, variety, and veracity). Veracity is an indication of data integrity and an organization’s ability to trust and use the data to make critical decisions. In addition, big data today is commonly referred to as 5V (volume, velocity, variety, veracity, and value), adding a value element that means the creation of new values through big data. As the demand for big data processing has increased, platforms for storing and analyzing it have also been developed. As a result, big data is now rapidly being applied across all scientific and industrial domains, including healthcare [12,13].
The source of medical big data includes electronic health record (EHR) data, various information stored in hospital information systems, including diagnosis, test results, prescription, and even structured medical records. Recent advances in diagnostic technology have produced big data, including diagnostic imaging, EEG, and genetic testing. The strength of big data research using data stored in the HIS or the national insurance data has increased statistical power compared to previous studies that used a limited sample to estimate the findings of a population. Using time series and digital imaging bid data, we can perform ML or DL to construct AI algorithms. Using time series big data such as EEG, we can predict seizures using DL techniques from multiple channels of intracranial EEG with 4096 data points/s over a couple of days. DL algorithms detect lesions from chest x-rays, brain computed tomography, and magnetic resonance imaging using image big data. Relevant issues in using big data for medical research include digitalization of data, unifying the data format, and data validation [14]. We will introduce the CDM analysis in epilepsy as a sample of HIS big data research.
2. Common data model
CDM is a data model that enables data owners to support observational health studies through a distributed research network while protecting patient health information by maintaining the data in the institution. The CDM standardizes different data types of each institution with the same structure and terminology, allowing research participants to share analytics codes and collect analysis results to enable scalable large-scale big data research. The standardization ensures that research methods and analytics can be systematically applied to generate meaningfully comparable and reproducible results [15]. By converting EHR data and claims data to CDM, interest and opportunities for generating reliable and reproducible evidence from real-world health data are increasing. There are several types of CDMs: Sentinel, i2b2, Clinical Data Interchange Standards Consortium Study Data Tabulation Model, National Patient-Centered Clinical Data Research Network, and Observational Medical Outcomes Partnership (OMOP) depending on the purpose of the data utilization and the leading institution [16-20]. OMOP CDM is differentiated in that it seeks to ensure semantic interoperability by adopting international standard terminology.
Observational Health Data Sciences and Informatics (OHDSI) maintains OMOP CDM as an open international community and supports pharmacovigilance research, clinical characterization, population-level estimation, and patient-level prediction studies by collaboratively developing data standards, research methodologies, and open-source analytic tools that can generate scientific evidence from observational health data [20-23]. More than 200 institutions worldwide have been converting EHR or claims data to OMOP CDM and are registered with the OHDSI data network [24].
CDM standardization has the advantage that reproducible studies can be performed using standardized analysis methodology from the federated data network. Especially for rare diseases, statistical power can be acquired using multicenter patient data. In CDM-based AI research, the possibility of model generalization can be confirmed by evaluating the external validation of the data of another institution for a predictive model learned from data from one institution. In addition, federated learning can be utilized for distributed data networks.
The limitation of CDM is that it requires a lot of time and manpower to build and intensive cooperation in various fields such as domain experts, vocabulary experts, software developers, data scientists, and time-consuming code mapping for vocabulary standardization. Another challenge is that there may be differences in the granularity and vocabulary mapping for each institution depending on the level of continuous maintenance and management according to the continuous upgrade of CDM versions and vocabularies. In particular, there is concern that the vocabulary mapping method or mapping granularity for each institution may differ, so that the mapping process can cause information loss and may affect the data analysis results [25]. Nevertheless, conversion to OMOP CDM will increase the opportunities for healthcare big data research in various diseases.
3. CDM research on epilepsy
A study from the OHDSI research network performed a big data analysis to address the recent issue of angioedema with the increased use of levetiracetam. They analyzed 10 databases to collect the data of 276,665 patients who used levetiracetam and 74,682 patients who used phenytoin to compare the incidence of angioedema. An increased incidence of angioedema with levetiracetam was seen in some databases; a meta-analysis of all databases did not show a significant difference compared to phenytoin [26]. This study is a good example considering the limited number of pediatric patients in the adult population. We can utilize CDM analysis for multiple institutions or regions to determine the actual raw data of the population. Our group recently performed CDM research to review ASM-related hematologic adverse drug reactions at our institution [27]. Our routine practice in epilepsy care is to perform regular blood tests to detect common hematologic adverse drug reaction at least annually. We built a query to identify patients who had regular outpatient visits, ASM prescriptions, and annual laboratory tests. To exclude the effects of polytherapy, the query was designed to select only the time periods when a single ASM was used.
Because the laboratory results are stored as a structured database in our LIS, we could easily collect the test results. From 1,344 pediatric epilepsy patients, we calculated the actual incidence of hyponatremia, leukocytopenia, thrombocytopenia, liver enzyme elevations, and thyroid function test abnormalities that occurred with oxcarbazepine, levetiracetam, valproic acid, topiramate, and lamotrigine monotherapy. We plan to apply the same query to other hospitals using the same CDM format to calculate regional and even national incidence. Another study using CDM was performed to apply the treatment pathways in ASM treatment. Many studies have analyzed large-scale treatment patterns in common chronic diseases such as hypertension, diabetes mellitus, and depression [28]. We also analyzed the treatment pathways in pediatric epilepsy patients [29]. We constructed a query to include pediatric epilepsy patients who underwent treatment for at least 2 years and attended regular follow-up and searched for their ASM treatment regimen. Sunburst diagrams of 1,192 patient treatment pathways are shown in Fig. 1. To demonstrate the applicability of this analysis in epilepsy care, we estimated the medication refractoriness based on our results. Since medication refractoriness is defined as failure of more than 2 most efficient ASM in a 6-month period, we applied the same definition and estimated a rate of 23.8% compared to various historical controls. We could even visualize annual ASM prescription pattern changes.
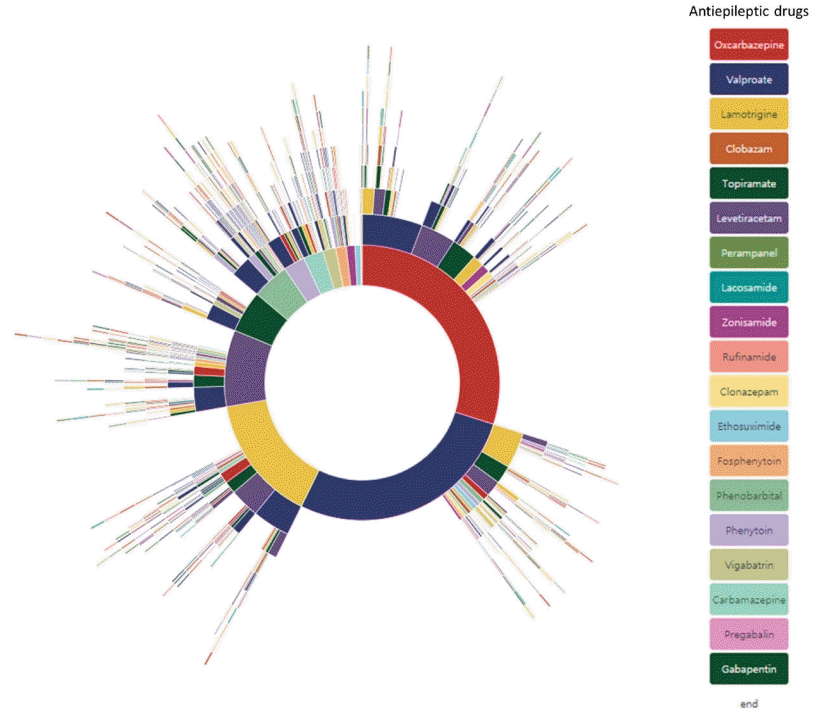
Treatment pathways of all 1,192 pediatric epilepsy patients. Specific antiseizure medications used and their sequence is shown in the sunburst plot.
Because the CDM database is completely anonymized and the data cannot be identified, it has great strength in personal information protection. However, there are critical issues in the successful CDM analysis. Outcome data and risk factors must be accurately available in the CDM database. Based on the research question and hypothesis, search and analysis queries must be accurate and adaptable for data retrieval. Careful consideration of these key issues can lead to successful CDM big data analysis.
AI in epilepsy electrophysiology
1. AI and DL
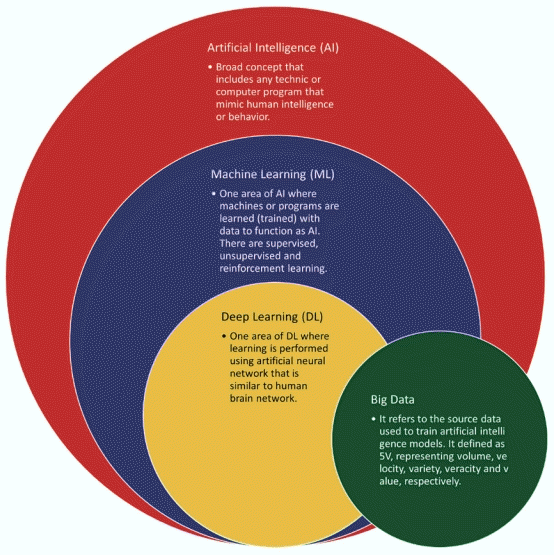
According to Gartner, AI meant applying advanced analysis and logic-based techniques, including ML, interpreting events, supporting and automating decisions, and taking actions. In other words, AI can be broadly classified into reasoning and learning systems. ML is a field of AI defined by Tom Mitchell in which a computer program is said to learn from experience E for some class of tasks T and performance measure P if its performance at tasks in T as measured by P improves with experience E. In general, there are 3 types of ML: supervised learning, unsupervised learning, and reinforcement learning. Supervised learning is the same as unsupervised learning methods based on input data, but the difference requires labeling of the input data. Reinforcement learning is learning how to map situations to actions to maximize a numerical reward signal [30]. In an actual ML system, a phenomenon called ‘overfitting’ may occur, model only memorizing the training data rather than finding a general predictive value [31]. Hence, ML is also an optimization problem that it attempts to solve using the correct objective function. The definitions of AI, ML, DL, and big data and their hierarchy and relationships are schematically shown in Fig. 2.
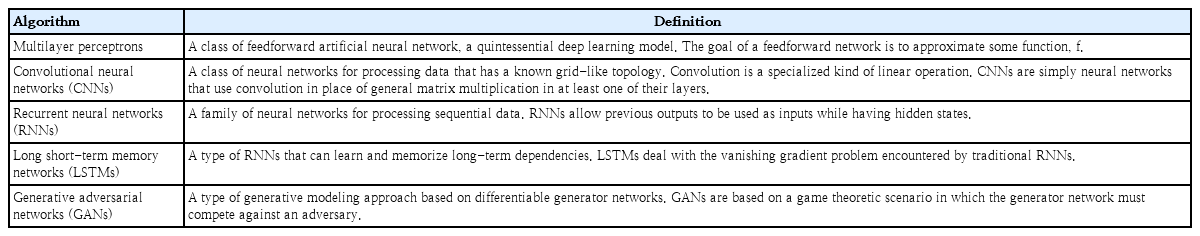
The origin of the term AI was reportedly introduced by John McCarthy in a workshop proposal held at Dartmouth University in 1956. AI research has since gained popularity in academia, but it went through 2 dark ages called “AI winter” in the 1970s and the late 1980s, respectively. With recent advancements in computer hardware, computing power has increased. The existing AI algorithm can be executed, and DL techniques that use more complicated arithmetic can emerge. DL methods are subfields of ML, a representation learning method with multiple levels of representation obtained by composing simple but nonlinear modules that each transform the representation at one level (starting with the raw input) into a representation at a higher but slightly more abstract level [32]. These methods have dramatically improved the state-of-the-art of speech recognition, visual object recognition, object detection, and many other domains such as drug discovery and genomics. For instance, in the field of image recognition, DL models showed fewer recognition errors than humans throughout the 2015 ImageNet Large Scale Visual Recognition Challenge competition [33]. Moreover, in 2015, Google’s DeepMind became the first computer program to beat a human professional Go player through AI called AlphaGo [34]. In healthcare areas, AI research is rapidly accelerating to demonstrate its potential applicability in many different domains of medicine [35]. Recently, explainable AI studies have also been actively underway to solve the black-box problem, known as the structural limitations of DL models [36-38]. We will present examples of AI application to EEG research aimed at spike detection, seizure detection, and seizure forecasting. To capture the intrinsic spatiotemporal dynamics of EEG, convolutional neural networks (CNNs) and long short-term memory (LSTM) networks, a specific type of recurrent neural network, have been widely suggested to handle detection and forecasting problems. The definitions of the recent DL algorithms are described in Table 1.
2. Application of AI to EEG
1) Automated detection of interictal epileptiform discharges
Detecting interictal epileptiform discharges (IEDs) on the EEGs of patients with epilepsy is critical. The current practice of EEG interpretation is a visual analysis that significantly depends on reader skillfulness. Numerous IEDs vary greatly, and it is crucial that skillful interpreters have extensive knowledge and experience. Automatic IED detection can help clinicians detect IEDs easily and precisely. It is helpful for quantifying the number of IEDs for further clinical care and research.
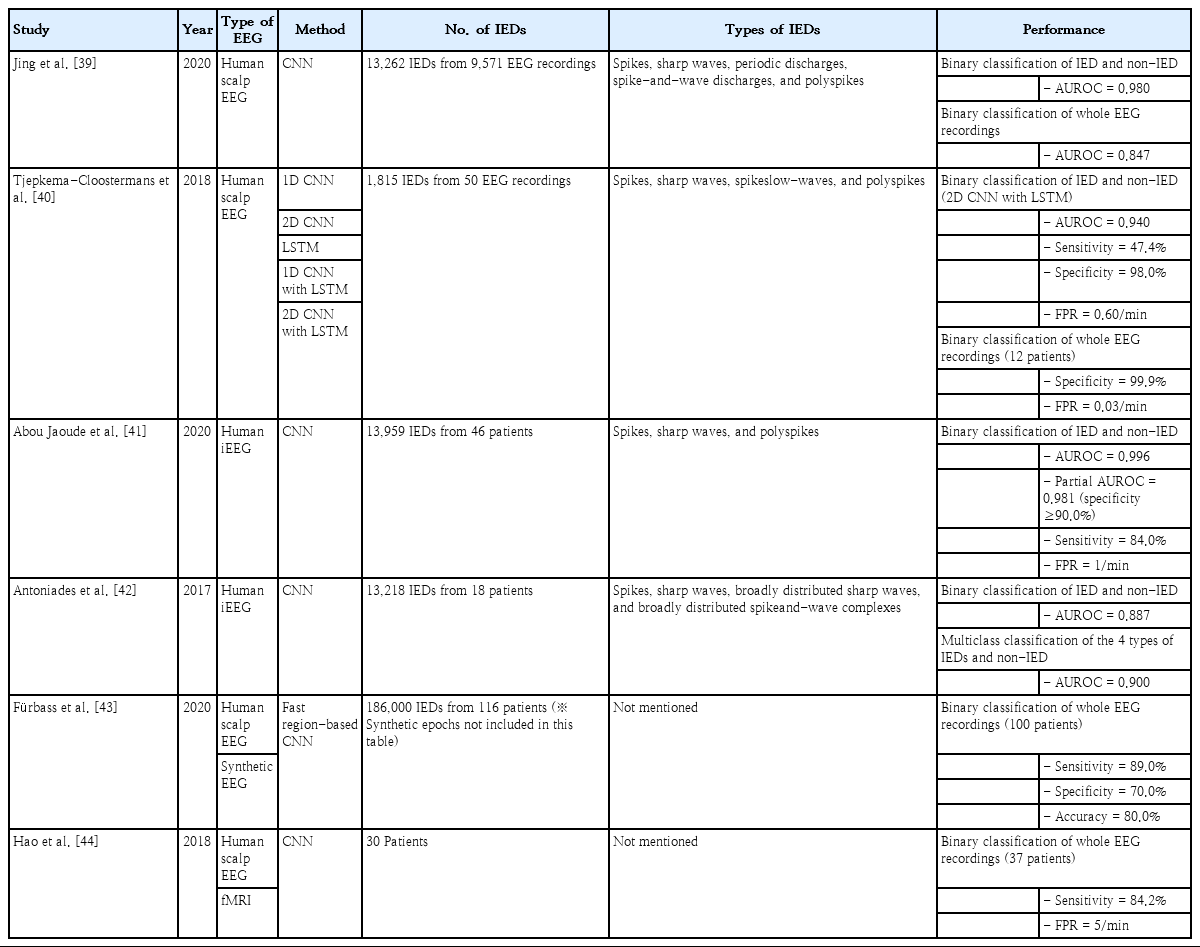
A recent study adopted a CNN to detect IEDs on human scalp EEG recordings. They implemented a CNN-based classification model, called SpikeNet, using 13,262 IEDs annotated by clinicians from 9571 EEG recordings to automatically detect IEDs, including spikes, sharp waves, periodic discharges, spike-and-wave discharges, and polyspikes. They achieved an area under the receiver operating characteristic curve (AUROC) of 0.980 for the binary classification of IEDs and non-IEDs, which surpassed both expert-level interpretation and industry-level performance (0.882) based on commercial software. In addition, they achieved an AUROC of 0.847 for another binary classification of whole EEG recordings with IEDs (IED-positive) and those without IEDs (IED-negative) [39].
A study that used both CNN and LSTM to detect IEDs on human scalp EEG recordings implemented classification models based on 1-dimensional (1D) CNN, 2-dimensional (2D) CNN, LSTM, 1D CNN with LSTM, and 2D CNN with LSTM using 1,815 IEDs annotated by clinicians from 50 EEG recordings to automatically detect IEDs, including spikes, sharp waves, spike-slow-waves, and polyspikes. They achieved an AUROC of 0.940 for the binary classification of IEDs and non-IEDs using the 2D CNN with LSTM, which outperformed the other models. The 2D CNN with LSTM model showed sensitivity, specificity, and a false-positive rate (FPR) of 47.4%, 98.0%, and 0.60/min, respectively. Moreover, it showed a specificity of 99.9% with an FPR of 0.03/min for additional validation data of 12 normal EEG recordings [40].
Abou Jaoude et al. [41] adopted a CNN to detect IEDs on human intracranial EEG recordings. They implemented a CNN-based classification model using 13,959 IEDs annotated by clinicians from 46 patients to automatically detect IEDs, including spikes, sharp waves, and polyspikes. They achieved an AUROC of 0.996 for the binary classification of IEDs and non-IEDs. In addition, they showed a partial AUROC of 0.981 with a specificity higher than 90.0% and a sensitivity of 84.0% with an FPR of 1/min. Another study that used a CNN-based classification model to detect IEDs in human intracranial EEG recordings used 13,218 IEDs annotated by clinicians from 18 patients to automatically detect IEDs, including spikes, sharp waves, broadly distributed sharp waves, and broadly distributed spike-and-wave complexes. They achieved an AUROC of 0.900 for the multiclass classification of the 4 types of IEDs and non-IEDs and 0.887 for the binary classification of IEDs and non-IEDs [42]. In addition, Fürbass et al. [43] proposed a semisupervised approach based on a fast region-based CNN to detect IEDs in human scalp EEG recordings with a sensitivity of 89.0% and a specificity of 70.0%, while Hao et al. [44] proposed a semiautomatic IED detector based on CNN using EEG-functional magnetic resonance imaging recordings with a sensitivity of 84.2%.
As shown above, many detectors and algorithms have been developed to detect IEDs. However, they are not widely used in the clinical setting. There are different IEDs, such as focal and generalized discharges. The distribution of the focal discharges varies widely. These issues must be solved to be widely used in practice. We are currently planning to develop novel DL-based automated IED detectors in terms of disease specificity and IED complexity. The detectors are expected to be applied to specific types of epilepsy syndromes, showing typical morphological characteristics of IEDs in clinical diagnosis. In addition, it is feasible to detect various complicated IEDs beyond the findings of previous studies, such as prolonged abnormal EEG activities including multiple IEDs through methodological modifications of well-known DL approaches.
2) Seizure detection
Automated seizure detectors can be used in continuous monitoring, such as in neuro-intensive care settings and video-EEG monitoring. They also target seizure detection using wearable devices with a reduced number of electrodes. Automated seizure detection with increased accuracy provides precise information regarding seizure counts for medication adjustment. They can be ideally used to activate the emergency medical system in cases of urgent situations such as status epilepticus.
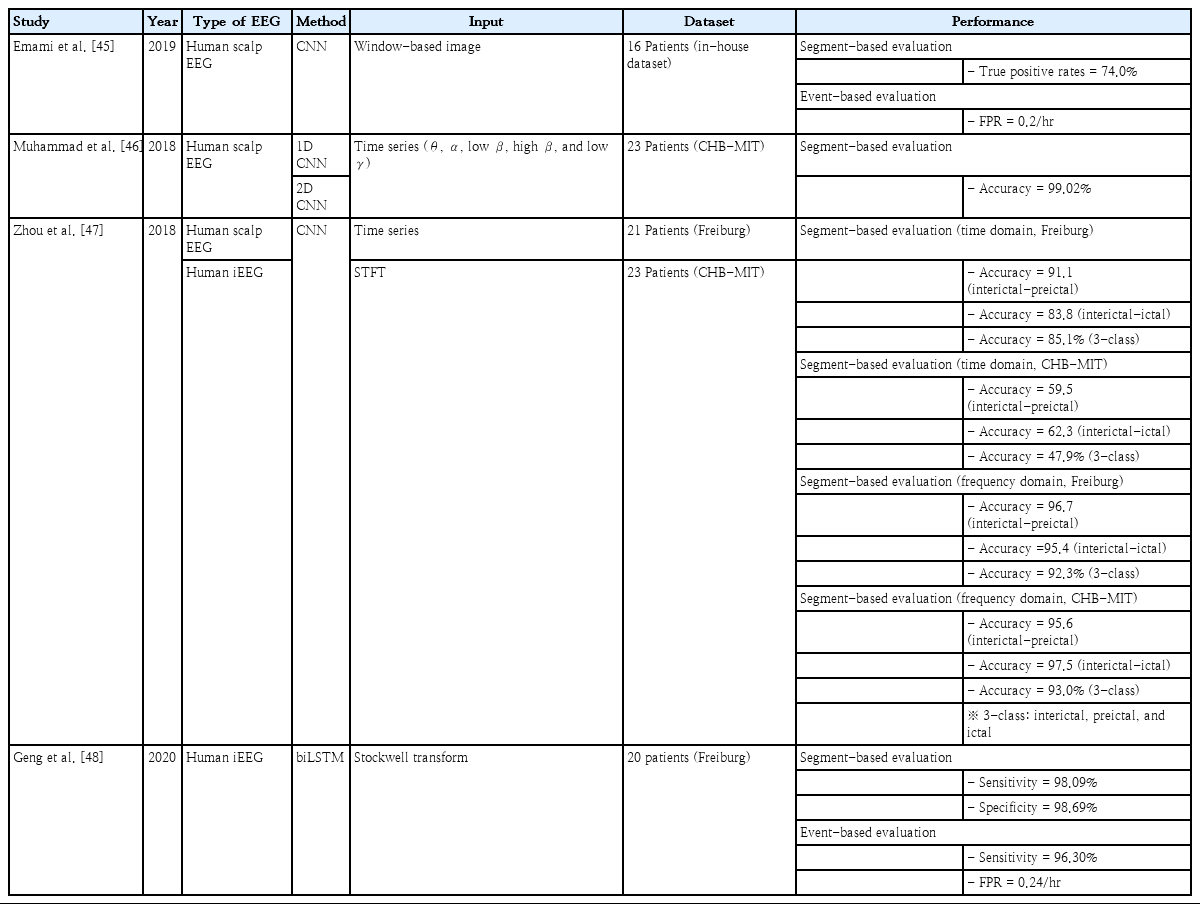
Emami et al. [45] adopted a CNN to detect seizures on human scalp EEG recordings. They implemented CNN-based classification models using their in-house dataset, including the EEG recordings of 16 patients. The multichannel EEG time series were converted into window-based images. A series of EEG images were fed into the models as input data to classify the seizure and nonseizure EEG segments. They achieved a median true positive rate of 74.0% with a 1-second time window in image-based seizure detection, and an FPR of 0.2/hr in the event-based evaluation for the detection of seizures immediately after onset. Their true positive rates were better than those of commercially available software (20.0%–31.0%), whereas their FPR was higher than that of the software.
Muhammad et al. [46] adopted a CNN to detect seizures on human scalp EEG recordings. They implemented classification models based on both 1D and 2D CNN using the Children’s Hospital Boston - the Massachusetts Institute of Technology (CHB-MIT) dataset, including EEG recordings of 23 patients. Band-limited EEG time series were fed into the models as input data. They used autoencoders to reduce the dimensionality of the features extracted by CNN layers to propose mobile multimedia healthcare systems. They achieved an accuracy of 99.02% for the binary classification of seizure and nonseizure segments.
Zhou et al. [47] adopted a CNN to detect seizures on the human scalp and intracranial EEG recordings. They implemented CNN-based classification models using time domain (EEG time series) and frequency domain (Fourier transform-based power spectral density) with the Freiburg dataset including EEG recordings of 21 patients and the CHB-MIT dataset including the EEG recordings of 23 patients. The CNN-based classifier was evaluated for 2 binary classifications (interictal-preictal and interictal-ictal states) and one 3-class classification (interictal, preictal, and ictal). Regarding the time-domain classification, they achieved an accuracy of 91.1%, 83.8%, and 85.1% in the Freiburg dataset versus 59.5%, 62.3%, and 47.9% in the CHB-MIT dataset for the 3 evaluations. Regarding the frequency domain classification, they achieved an accuracy of 96.7%, 95.4%, and 92.3% in the Freiburg dataset versus 95.6%, 97.5%, and 93.0% in the CHB-MIT dataset for the 3 evaluations, representing a pivotal role of the frequency information in seizure detection.
Geng et al. [48] adopted a bidirectional LSTM to detect seizures on human intracranial EEG recordings. They implemented the LSTM-based classification models using the 4-second time-frequency data transformed by the Stockwell transform with the Freiburg Hospital dataset, including the EEG recordings of 20 patients. They achieved a sensitivity of 98.09% and a specificity of 98.69% in segment-based evaluation, and a sensitivity of 96.30% with an FPR of 0.24/hr in event-based evaluation.
3) Seizure forecasting
Seizure forecasting is a highly challenging area of big data and AI research in the field of epilepsy. It is helpful for patients with intractable epilepsy whose seizures are not controlled by various ASMs. If we can accurately recognize upcoming seizures in less than a minute, we can prevent injuries that result from falls or body stiffening. Rescue medications such as rectal, intranasal, buccal, or sublingual benzodiazepines can prevent seizures if they can be forecasted a few minutes before onset. Although there are problems with false alarms, the consequences of unpredictable seizures are more detrimental to patients and their caregivers. The status of seizure forecasting research is just the beginning phase, and many researchers are currently working to develop an algorithm or system to predict seizures.
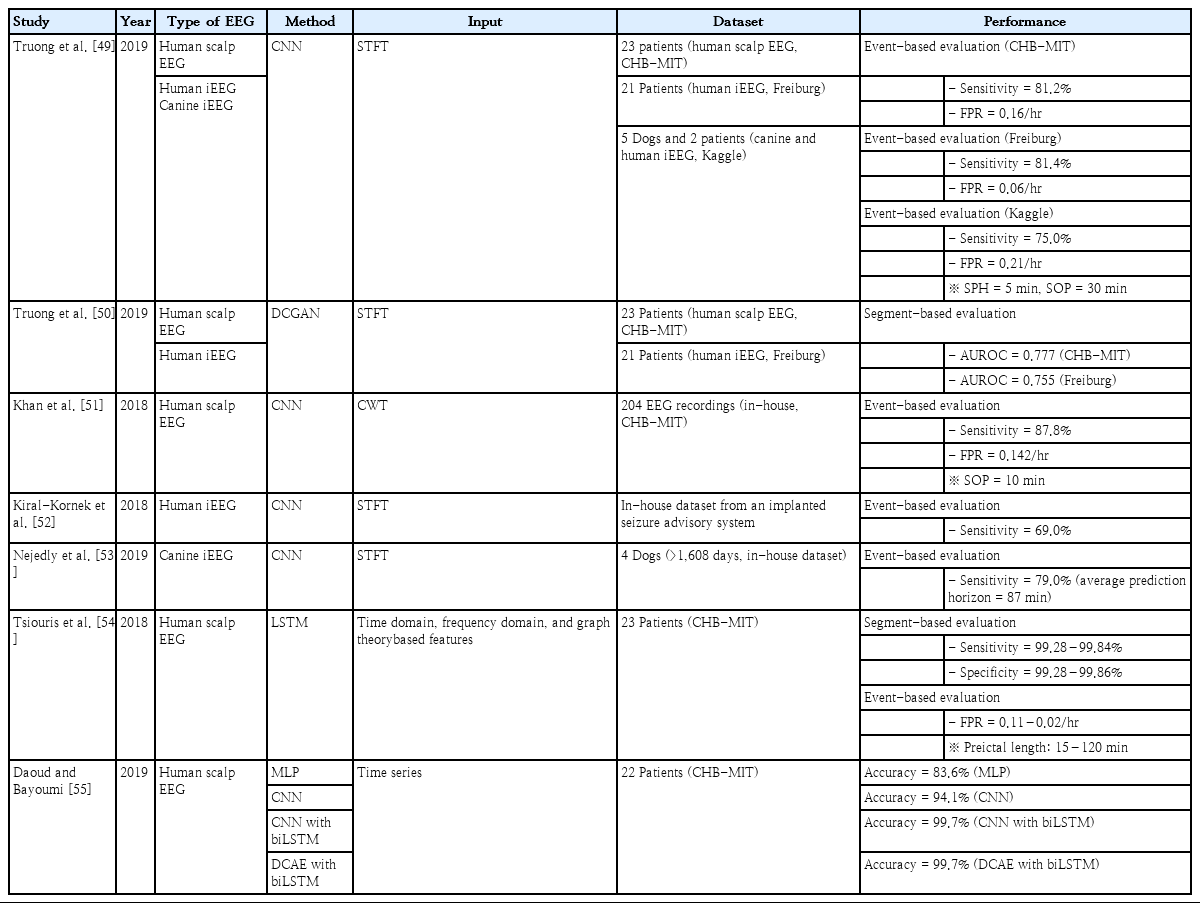
Truong et al. [49] adopted a CNN to predict seizures in human scalp intracranial EEG recordings and canine intracranial EEG recordings. They implemented CNN-based prediction systems using 30-second time-frequency EEG data transformed by the short-time Fourier transform (STFT) with publicly available datasets, including 23 patients from the CHB-MIT dataset (human scalp EEG), 21 patients from the Freiburg Hospital dataset (human intracranial EEG), and 5 dogs and 2 patients from the American Epilepsy Society Seizure Prediction Challenge (Kaggle) dataset. In the event-based evaluation for the prediction of upcoming seizures, they achieved a sensitivity of 81.2%, 81.4%, and 75.0% and an FPR of 0.16/hr, 0.06/hr, and 0.21/hr with a 5-minute seizure prediction horizon and a 30-minute seizure occurrence period (SOP) in the CHB-MIT, Freiburg Hospital, and Kaggle datasets, respectively. They exploited deep convolutional generative adversarial networks (DCGANs) as an unsupervised feature extraction technique to minimize laborious and time-consuming labeling tasks. They implemented DCGAN-based classification models using the time-frequency EEG data under similar experimental conditions and achieved AUROCs of 0.777 and 0.755 for the binary classification of seizure and nonseizure segments in the CHBMIT and Freiburg datasets, respectively [50].
Khan et al. [51] adopted a CNN to anticipate the occurrence of seizures on human scalp EEG recordings. They implemented CNN-based prediction systems using EEG wavelet tensors transformed by the continuous wavelet transform with 204 EEG recordings from their in-house and CHB-MIT datasets. An optimal preictal length of 10 minutes was estimated based on the probability distributions of interictal and preictal features. They achieved a sensitivity of 87.8% and an FPR of 0.142/hr with a 10-minute SOP in the event-based evaluation.
Kiral-Kornek et al. [52] adopted a CNN to predict seizures on human intracranial EEG recordings. They implemented CNN-based prediction systems using time-frequency EEG data transformed by STFT with their in-house dataset collected by an implanted seizure advisory system [14]. They achieved a sensitivity of 69.0% in the event-based evaluation and demonstrated the feasibility of the deployment of their prediction models onto an ultra-low power neuromorphic chip in a wearable device.
Nejedly et al. [53] adopted a CNN to predict seizures on canine intracranial EEG recordings. They implemented CNN-based prediction systems using 30-second time-frequency EEG data transformed by STFT with their in-house dataset, including 75 seizures collected over 1,608 days in 4 dogs. They achieved a sensitivity of 79.0% with an average prediction horizon of 87 minutes in event-based evaluations.
Tsiouris et al. [54] adopted LSTM to predict seizures on human scalp EEG recordings. They extracted the time domain, frequency domain, and graph theory-based features from the EEG recordings of 23 patients from the CHB-MIT dataset as input data for the LSTM. In the segment-based evaluation, they achieved a sensitivity and specificity of 99.28%–99.84% and 99.28%–99.86%, respectively. In the event-based evaluation, they achieved an FPR of 0.11–0.02/hr at a 15- to 120-minute preictal duration. They demonstrated an advantage of the LSTM-based models for seizure prediction versus conventional ML-based models, suggesting that LSTM is a suitable approach to handle chaotic EEG characteristics.
Daoud and Bayoumi [55] proposed 4 different ML-based models to compare their performance for the binary classification of interictal and preictal states. The multichannel EEG time series were fed into the classification models as input data. They showed an accuracy of 83.6%, 94.1%, 99.7%, and 99.7% for multilayer perceptron (MLP), CNN with MLP, CNN with bidirectional LSTM, and deep convolutional autoencoder (DCAE) with bidirectional LSTM, respectively. They also suggested a channelselection algorithm based on entropy to enhance the model efficiency. The training time of the DCAE with the bidirectional LSTM model was reduced to almost half (from 4.25 minutes to 2.20 minutes) using the channel-selection algorithm.
Preictal states are known to exhibit noticeable electrophysiological characteristics that are distinguishable from interictal states [56-58]. Therefore, proper selection of the length of preictal states has been a crucial factor for seizure forecasting despite some previous studies solving this issue [51,59]. Hardware implementation of DL-based seizure prediction models is expected to facilitate their clinical application similar to responsive neurostimulation systems [60]. In this regard, we investigated interictal-preictal classification performance by varying the preictal length, number of electrodes, and sampling frequency to explore practical conditions to sufficiently discriminate preictal and interictal states in terms of EEG dynamics and data efficiency.
We implemented classification models based on 2D CNN using the 30-second time-frequency EEG data transformed by STFT with our in-house dataset, including intracranial EEG recordings of 9 patients with focal cortical dysplasia type II. We observed 3 main findings. The 5-minute preictal length showed the best interictal-preictal classification performance (higher than 13% accuracy compared to the 120-minute preictal length). Four electrodes showed considerably high accuracy, similar to all electrodes (approximately 1% reduction) with a 5-minute preictal length. The performance variability was substantially weak at the 128–512 Hz sampling frequencies. Regarding patient-specific evaluations, the preictal length affected performance enhancement (higher than 28% for 3 patients) stronger than the other factors, suggesting the importance of pre-examinations for optimal preictal lengths to devise clinically feasible seizure prediction systems [61].
Recent DL-based studies of IED detection, seizure detection, and seizure forecasting are summarized in Tables 2–4. They achieved 0.887–0.996 AUROCs for automated IED detection; 62.3%–99.0% accuracy for interictal classification in seizure detection; and 75.0%–87.8% sensitivity with a 0.06–0.21/hr FPR for seizure forecasting.
Suggestions for clinicians
There is no doubt that big data and AI have changed our everyday lives. They have also started to change the field of medical research and open a new arena. In addition to previous medical studies that has greatly improved patient well-being and treatment, we hope that new tools such as big data analysis and AI application will answer unsolved questions and broaden the knowledge of human diseases. New methodologies are not suitable for previous research. However, they complement previous tools. The vague fear that AI will replace humans in various aspects of society is also present in medicine. The application of AI in medical research and practice is aimed at improving the performance or outcome in addition to human effort. The algorithms are designed and programmed by humans and modified by human researchers and developers to enhance their functions. The transition to digital healthcare has generated and accumulated different kinds of big data. The development of wearable devices to monitor blood pressure, electrocardiography, glucose levels, EEGs, and many other biosignals produces other forms of big data.
To adapt to current changes, clinicians should be aware of the changes that have been around us for the last few decades. We should have basic knowledge of big data and AI to help us understand and interpret related research accordingly. It is also valuable to understand the structure and design of AI algorithms applied in clinical practice. Another role of clinicians is to provide research questions and insight to researchers in these fields. Although the number of clinicians actively involved in this research is increasing, most studies have been performed with 2 different clinicians and AI researchers. Therefore, it is critical that clinicians provide the correct hypothesis, adequate data, and clinically meaningful interpretations.
One of the most critical roles of clinicians in big data and AI research is to highlight research questions or unmet needs in practice where these new methods can provide breakthrough changes. Finally, constructing a structured database of medical records is essential. There are still significant limitations in utilizing medical records for big data analysis and training for ML algorithms. We can use text mining or natural language processing to retrieve relevant information and construct a structured database. Still, it is another area of research being tested in the field. As we use structured data entry in clinical trials, there has been an effort to create common data elements (CDE) for research on neurological disorders from the National Institute of Neurological Disorders and Stroke [62]. Based on this movement, researchers created CDE for clinical practice in epilepsy for its later research use [63]. This is a good example of a structured database of clinical information for big data analysis and AI applications.
Notes
Conflicts of interest
No potential conflict of interest relevant to this article was reported.
Funding
This study received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.