Neonatal risk factors associated with autism spectrum disorders: an umbrella review
Article information
Abstract
Background
Autism spectrum disorder (ASD) is a neurodevelopmental disorder characterized by severe social communication deficits and stereotypical repetitive behaviors.
Purpose
This umbrella review assessed neonatal risk factors associated with ASD using meta-analyses and systematic reviews.
Methods
We conducted a systematic search of international databases including PubMed, Scopus, and Web of Science for studies published through April 2022 utilizing pertinent keywords. A random-effects model was used to calculate the odds ratio (OR) and 95% confidence interval (CI). Substantial heterogeneity was considered at values of I2≥50%. A quality assessment of the included studies was performed using the A MeaSurement Tool to Assess Systematic Reviews (AMSTAR2) checklist.
Results
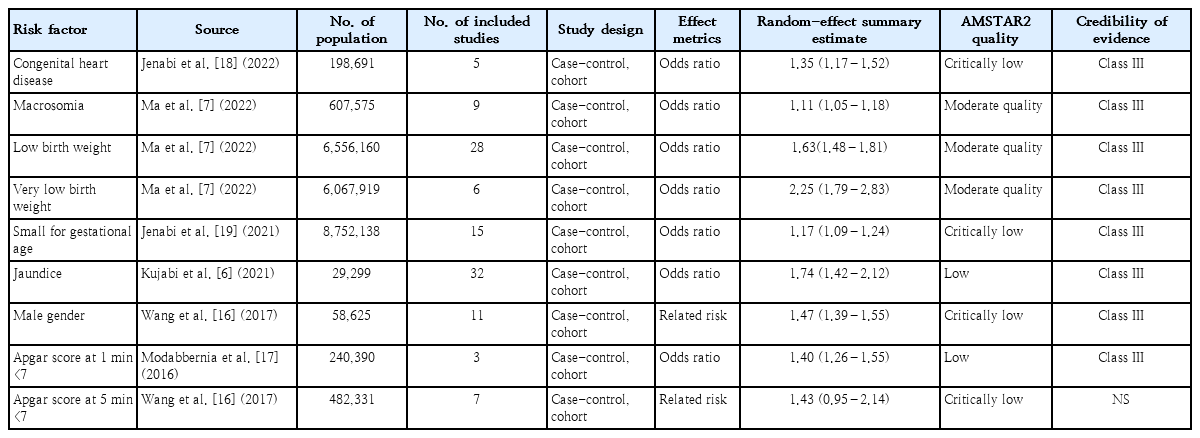
A total of 207,221 children with ASD and 22,993,128 neurotypical children were included. Six meta-analyses were included in this umbrella review. The factors of congenital heart disease (OR, 1.35; 95% CI, 1.17–1.52), macrosomia (OR, 1.11; 95% CI, 1.05–1.18), low birth weight (OR, 1.63; 95% CI, 1.48–1.81), very low birth weight (OR, 2.25; 95% CI, 1.79–2.83), small for gestational age (OR,1.17; 95% CI, 1.09–1.24), jaundice (OR, 1.74; 95% CI, 1.42–2.12), male sex (OR, 1.47; 95% CI, 1.39–1.55) and 1-minuteApgar score <7 (OR, 1.40; 95% CI, 1.26–1.55) were graded as suggestive evidence (class III). Only 3 studies reported heterogeneity (I2<50%). Based on the AMSTAR2 analysis, the methodological quality was critically low in 3 meta- analyses, low in 2, and moderate in 1.
Conclusion
Based on these results, clinicians should consider the risk factors for ASD and screen children in clinics.
Key message
Question: What are the neonatal risk factors for autism spectrum disorder (ASD)?
Findings: Significant effect sizes were observed for congenital heart disease (odds ratio [OR], 1.35), macrosomia (OR, 1.11), low birth weight (OR, 1.63), very low birth weight (OR, 2.25), small for gestational age (OR, 1.17), jaundice (OR, 1.74), male sex (OR, 1.47), and Apgar score (OR, 1.40).
Meaning: These factors were identified as risk factors for ASD.
Introduction
Autism spectrum disorder (ASD) is a neurodevelopmental disease that limits social activities, and these individuals have difficulty with verbal and nonverbal communication, social interactions, and adaptive skills. ASD is often associated with the impaired cognitive function of learning and sensory processing and is usually diagnosed in childhood [1]. The prevalence of this disease is increasing worldwide [2]. The prevalence of autism in the United States is 1 in 36 children and is higher in boys (1 in 23 boys) [3].
The main cause of this disease has not been entirely determined; however, there is strong evidence in favour of genetic factors, although environmental factors also play a role [4]. Various studies have been performed on the risk factors for ASD; these factors include risk factors related to maternal and pregnancy, such as preterm labor, maternal age, bleeding during pregnancy, and drug use during pregnancy [5], and neonatal risk factors such as low weight, jaundice, congenital heart disease (CHD), low birth weight (LBW), very LBW (VLBW) and small size for gestational age (SGA) [6-9].
The presence of cognitive and learning challenges in individuals with ASD, along with the consequential negative effects on their social interactions with the surrounding world, and the escalating costs to the health system, has prompted researchers to dedicate significant attention to this disorder. Researchers are actively investigating the various factors influencing the development and impact of the disease.
To the best of our knowledge, no umbrella reviews have been conducted to evaluate neonatal risk factors linked to ASD through meta-analyses and systematic reviews. Hence, the current umbrella review aims to assess neonatal risk factors associated with ASD, drawing on findings from existing meta-analyses and systematic reviews.
Methods
We performed the current umbrella systematic review according to the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guideline [10]. Moreover, we registered the study protocol at the International Prospective Register of Systematic Reviews (PROSPERO) with register number CRD42022326480, Date: 10/05/2022 and URL https://www.crd.york.ac.uk/PROSPERO/#record Details.
The Ethics Committee of the Hamadan University of Medical Sciences approved the protocol of this study (IR.UMSHA.REC.1401.153.). The methods were performed in accordance with the ethical standards as laid down in the Declaration of Helsinki and its later amendments or comparable ethical standards.
1. Inclusion and exclusion criteria
The present study included all systematic reviews with observational studies (case-control, cohort, and cross-sectional design). Conference abstracts, letter to the editors, and original articles were excluded. We exclusively considered meta-analyses pertinent to neonatal risk factors associated with ASD. Inclusion in the umbrella study was contingent on a meta-analysis explicitly providing the essential information for analyses; otherwise, the study was excluded. Moreover, animal studies were also excluded from our study selection. If there are 2 or more meta-analyses related to a factor, we include the meta-analysis with the largest number of studies. There was no language restrictions in searching strategy.
2. Searching strategy
A combination of search terms related to the effect of neonatal risk factors associated with ASD was used to retrieve eligible systematic reviews in the current umbrella study. We searched the 3 major international databases, including PubMed, Scopus, and Web of Sciences, from inception until April 10, 2022, without any restriction in language and time. The details regarding the search strategy have been provided in the supporting material (Supplementary Table 1).
3. Study selection
The searching procedure was done by 2 authors (EJ and SK) independently. After duplicate removal and assessing the articles' title, abstract, and full text, the eligible studies were screened. Any disagreements between authors were resolved through negotiation. In continued to find any possible missed study during the search in databases, the reference list of included studies was assessed carefully. The selection of eligible sources followed a PECO (Population, Exposure, Comparison, and Outcome) model. The population encompassed systematic reviews that centered on ASD in human samples. The exposure focused on neonatal risk factors. Comparison was conducted based on the acquired odds ratio (OR) or relevant risk (RR) ratio from the studies, and the outcomes were assessed in terms of the association between these risk factors and ASD.
4. Data extraction
Data extraction was performed by the 2 authors (EJ and SK) independently. Disagreement between authors was resolved by discussion with a third author (EA). The retrieved information from the studies included the first author name, publication year, population sample size, number of included studies in the meta-analysis, study design, related risk factors, the pooled effect size and related confidence interval (CI), and credibility of the evidence. The nominally significant P value for the largest study was extracted for each meta-analysis. Additionally, the quality of each study, the heterogeneity among studies, and the publication bias for each reviewed study were calculated and reported.
5. Quality assessment analysis
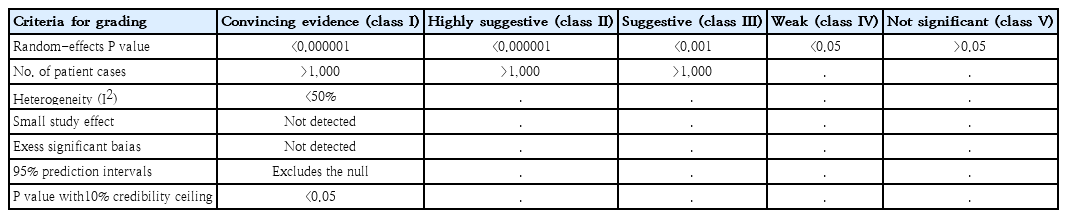
We used the “A MeaSurement Tool to Assess Systematic Reviews (AMSTAR2)” tool for assessing the quality of the included meta-analyses [11]. This process was done by 2 authors (EJ and SK) independently and any disagreement was resolved through negotiations between authors. We used AMSTAR2, which consists of 16 questions (Supplementary Table 2).
Each question is assigned a score of "yes," "partial yes," or "no." Among them, items 2, 4, 7, 9, 11, 13, and 15 are considered critical items. The cumulative score derived from these critical questions dictates the overall quality rating of the meta-analysis. This overall rating can be classified as high, moderate, low, or critically low, offering a comprehensive assessment of study quality and reliability.
6. Statistical analysis
All statistical analyses in this study were executed using R ver. 4.0.5 (R Foundation for Statistical Computing, Vienna, Austria). Various R packages, including Metafor, xlsx, ConfoundedMeta, Reporter, and epiR, were utilized. The included meta-analyses were reanalyzed based on the data supplied in the original papers, encompassing contingency tables, P values, and sample sizes. In instances of missing information, correspondence was initiated with the corresponding authors of the original papers to request the necessary data. If no response was received, the original paper with missing information was excluded from the reanalysis process.
During the reanalysis step, a random-effect model was employed for calculating pooled effect sizes and associated 95% CIs, as well as P values. The significance level for all statistical analyses was set at 0.05. Heterogeneity between studies was assessed using Cochrane Q test and the I2 statistic [12]. Substantial heterogeneity was considered when I2 was 50% or higher. Additionally, the prediction interval was calculated, providing an indication of the interval within which the estimate of the effect size by future studies might lie 95% of the time. The assessment of small study effects was conducted using Egger regression test [13]. This test indicates the potential bias in the estimated effect resulting from the larger treatment effects observed in smaller studies.
7. Sensitivity analysis
To assess the robustness of the meta-analyses to biases and unmeasured confounders, Mathur's method was employed as the sensitivity analysis approach [14]. Using this method, a bias factor and a paired confounding association strength were obtained for each meta-analysis.
Results
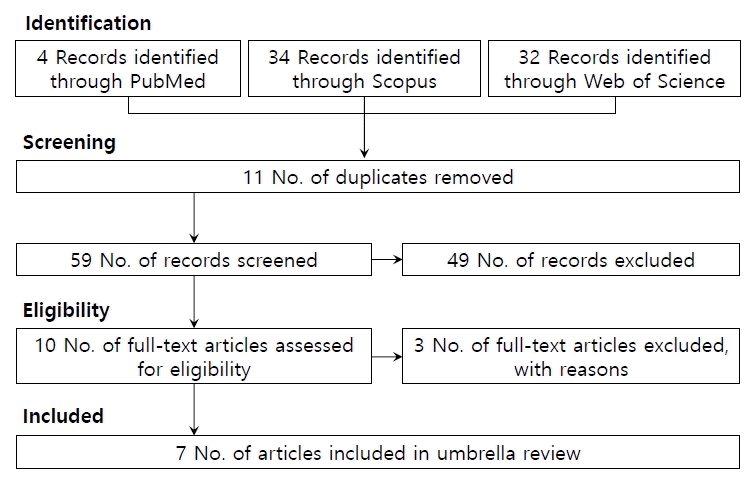
In the 3 databases, 70 articles were searched until April 17, 2022. We included 6 meta-analyses studies in the present umbrella review (Fig. 1). The detail the reasons for exclusion in screening was listed in the Supplementary Table 3. In total, 6 meta-analyses [6-8,15-17] included 9 studies with 207,221 children with ASD and 22,993,128 participants. Studies included in the present umbrella review had cohort and case-control designs (104 original, 26 cohort studies, and 78 case-control studies). In the present umbrella review, 8 risk factors found were CHD, macrosomia, LBW, VLBW, small for gestational age (SGA), jaundice, male gender, and Apgar score at 1 minute <7.
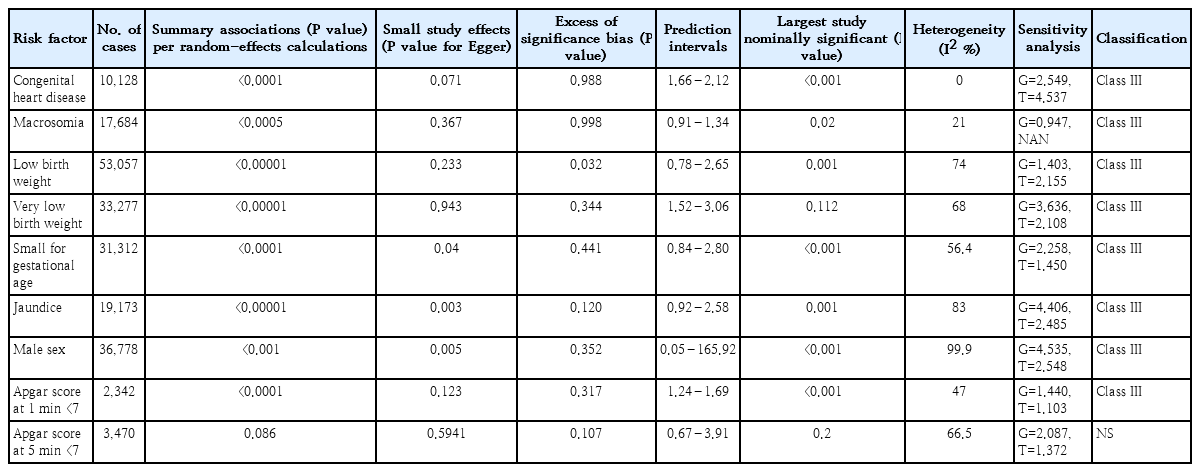
Out of the 9 associations in the umbrella review, the association of 8 analyses was statistically significant using the random-effects model, all studies had at least 1,000 children with ASD, 3 reported heterogeneity (I2) <50%, 4 had small study effects, and analyses had excess significance bias (Tables 2, 3).
The results of sensitivity analyses revealed that 4 meta-analyses (macrosomia, LBW, SGA, and Apgar score at 1 minute <7) were relatively sensitive to unmeasured confounding, with a bias factor less than 1.75 in each of their included studies based on previous studies [18,19]. These analyses indicated the potential to reduce the percentage of studies with a true OR greater than 1.1 to less than 20%. In contrast, the other 4 meta-analyses (CHD, VLBW, jaundice, and male gender) demonstrated relative robustness to unmeasured confounding, with a bias factor exceeding 1.90 in each of the included studies. This suggested the ability to reduce the percentage of studies with a true OR greater than 1.1 to less than 10%. Notably, Apgar score at 5 minutes <7 was not identified as a risk factor.
Eight risk factors of CHD (OR, 1.35; 95% CI, 1.17–1.52), macrosomia (OR, 1.11; 95% CI, 1.05–1.18) and Apgar score at 1 minute <7 (OR, 1.40; 95% CI, 1.26–1.55), LBW (OR, 1.63; 95% CI, 1.48–1.81), VLBW (OR, 2.25; 95% CI, 1.79–2.83), SGA (OR, 1.17; 95% CI, 1.09–1.24), jaundice (OR, 1.74; 95% CI, 1.42–2.12) and male gender (RR, 1.47; 95% CI, 1.39–1.55) were graded as suggestive evidence (class III). (Table 3). Apgar score at 5 minutes <7 (RR, 1.43; 95% CI, 0.95–2.14) was not identified as a risk factor. The methodological quality of the included meta-analyses, based on AMSTAR 2, was critically low in 3 meta-analyses , low in 2 meta-analyses and moderate in one meta-analysis (Table 2 and Supplementary Table 2).
Discussion
This is the first umbrella review that provides a robust synthesis of systematic reviews and meta-analyses regarding the neonatal risk factors of ASD. Overall, 8 risk factors including CHD, macrosomia, and Apgar score at 1 minute <7, LBW, VLBW, SGA, jaundice, male gender were supported by suggestive evidence (class III) and only Apgar score at 5 minutes <7 was nonsignificant. Despite the inclusion of meta-analyses with large sample sizes and a substantial number of cases (e.g., >1,000), none of the risk factors in these meta-analyses could attain a level of convincing or highly suggestive evidence. The failure to achieve a high level of evidence may be attributed to factors such as heterogeneity and potential biases [20].
Our results suggested that the small study effect may be a threat to the validity of meta-analyses of risk factors including SGA, jaundice, male gender, CHD, and ASD. This small study's effects can be explained by outcome reportig bias [21,22]. It means in the published studies evaluating these neonatal risk factors and adverse outcomes, there is a selective reporting of outcomes, so that odds of reporting associations for ASD in the main results of published studies may be lower than other outcomes. Another reason for small study effects may be due to clinical heterogeneity between ASD patients in large and small studies that evaluated the association between the aforementioned risk factors and ASD [22]. Our results provide clues for substantial between-study heterogeneity (>50%) among meta-analyses of the aforementioned risk factors and ASD. In our study, meta-analyses for CHD, macrosomia, VLBW, and Apgar scores comprised fewer than 10 studies. It's crucial to note that forming judgments solely based on statistical tests may be misleading. It has been argued that results from statistical tests for small study effect, excess significance tests, and heterogeneity can be biased when meta-analyses include less than 10 studies. Therefore, caution should be exercised in drawing conclusions solely from statistical analyses in such cases [23,24].
All associations between neonatal risk factors and ASD were consistent in strength and direction.It may be because there are strong interrelationships and plausible causal links in nature among most of these risk factors. Macrosomic infants are more often male and with lower Apgar scores and may experience hyperbilirubinemia [25,26]. Moreover, infants diagnosed with CHD are often SGA and LBW [27,28]. Hence, it can be inferred that complex interactions among these risk factors are important components in the causal path to ASD.
The random-effect summary estimate for the effect of SGA and macrosomia on ASD tends toward a null value. As shown in the sensitivity analysis, the results of the corresponding meta-analyses of these 2 risk factors may be sensitive to unmeasured confounding. It can be inferred that this unmeasured confounding is due to ignoring negative confounders. In studies that evaluated the association between SGA and macrosomia and ASD, a lack of control for negative confounders leads to the dilution of the effective measures. On the other hand, shrinkage of the summary estimates towards 1 may be a sign of the misclassification bias. For example, estimates of gestational age are prone to misclassification and it can lead to biased estimates for the effect of SGA on outcomes [29].
The association between most neonatal risk factors and ASD was investigated through case-control and cohort studies, relying on meta-analyses of ORs. While OR is a commonly used and preferred effect measure in meta-analysis, it's worth noting that using OR in cohort studies instead of risk ratio can be a subject of controversy in certain scenarios [30]. However, within the context of this study, the estimated ORs for the association between neonatal risk factors and ASD can offer a reasonable approximation of the relative measure of associations. This is particularly valid as the ORs are not significantly deviating from 1, and ASD is not a common outcome across different levels of the studied neonatal risk factors.
Our umbrella review is subject to several limitations. Most of the included meta-analyses in this review were critically low and low quality. In addition, a significant portion of the included systematic reviews and meta-analyses is susceptible to paper selection bias and publication bias since they did not make efforts to retrieve grey literature. Additionally, heterogeneity and potential biases, including small study effects in the meta-analyses, hinder the attainment of a high level of evidence.
In conclusion, this umbrella review of meta-analyses identified 8 significant neonatal risk factors associated with ASD. Our results showed 8 risk factors with suggestive evidence including CHD, macrosomia, and Apgar score at 1 minute <7, LBW, VLBW, SGA, jaundice, and male gender. Furthermore, future large-scale and prospective research could be directed towards assessing the impact of specific neonatal risk factors, such as CHD, Apgar score, and macrosomia, on the risk of ASD.It is important to note that the observed associations in umbrella reviews do not necessarily imply causation.
Supplementary materials
Supplementary Tables 1–3 can be found via https://doi.org/10.3345/cep.2024.00136.
Supplementary Table 1. Strategy search
cep-2024-00136-Supplementary-Table-1.pdfSupplementary Table 2. Qquality of studies based on AMSTAR2 items
cep-2024-00136-Supplementary-Table-2.pdfSupplementary Table 3. Excluded references with reasons
cep-2024-00136-Supplementary-Table-3.pdfNotes
Conflicts of interest
No potential conflict of interest relevant to this article was reported.
Funding
The protocol of this study was supported by Hamadan University of Medical Sciences with code1401022 71363
Author Contribution
Conceptualization: EJ, EA, SK, AS; Data curation: EA, EJ, EA, AS; Methodology: EJ, SB, AS; Software: EJ, ZS; Writing—original draft: EA, EJ, AS; Writing review, and editing: All authors.