All issues > Volume 68(1); 2025
Carbapenem resistance in gram-negative pathogens in an Iranian hospital: high prevalence of OXA-type carbapenemase genes
- Corresponding author: Shima Mahmoudi, PhD. Biotechnology Centre, Silesian University of Technology, 44-100 Gliwice, Poland Email: shima.mahmoudi@polsl.pl, sh-mahmoudi@sina.tums.ac.ir
- Received December 26, 2023 Revised June 11, 2024 Accepted June 24, 2024
- Abstract
-
- Background
- Background
- The widespread dissemination of carbapenem-resistant gram-negative bacteria poses a significant threat to global public health.
- Purpose
- Purpose
- This study aimed to investigate the prevalence of carbapenem resistance in gram-negative bacteria isolated from patients at the Children's Medical Center Hospital, Tehran, Iran, to understand the molecular mechanisms underlying this resistance.
- Methods
- Methods
- During the period spanning from June 2019 to June 2020, 777 gram-negative bacterial strains were isolated. Antibiotic susceptibility testing was performed according to Clinical and Laboratory Standards Institute. Polymerase chain reaction was used to detect carbapenem resistance genes including bla OXA23, bla OXA24, bla OXA48, bla OXA51, bla OXA58, bla OXA143, bla KPC, bla IMP, bla VIM, and bla NDM.
- Results
- Results
- Among the total bacterial isolates, 141 (18.1%) exhibited carbapenem resistance. Escherichia coli was the most prevalent (57.4%), followed by Klebsiella pneumoniae (11.3%), and Acinetobacter baumannii (10.6%). Other notable contributors included Enterobacter spp. (5.7%), Salmonella spp. (3.5%), and Stenotrophomonas maltophilia (2.8%). Citrobacter spp., Proteus mirabilis, and Pseudomonas aeruginosa contributed to the distributions of 2, 1, and 3 isolates, respectively. Notably, bla OXA48 showed the highest prevalence (33%), followed by bla OXA143 and bla OXA5 8 (27% and 24%, respectively). In addition, bla OXA24 was present in 11% of the total isolates, bla OXA23 in 10%, and bla NDM in 10%, whereas bla KPC, bla VIM, and bla IMP were not detected.
- Conclusion
- Conclusion
- Our study highlights the prevalence of carbapenemase-producing gram-negative isolates among pediatric patients. Notable resistance patterns, especially in K. pneumoniae and E. coli, underline the urgent need for proactive interventions, including appropriate antibiotic prescription practices and strengthening of antibiotic stewardship programs.
- Introduction
- Introduction
The widespread dissemination of carbapenem-resistant gram-negative bacteria poses a significant threat to public health globally [1,2]. The World Health Organization as identified carbapenem-resistant Enterobacteriaceae, carbapenem-resistant Pseudomonas aeruginosa, and carbapenem-resistant Acinetobacter baumannii as urgent concerns, guiding research and development efforts for new antibiotics [3].Carbapenemases represent a diverse group of enzymes that confer resistance to carbapenem antibiotics, posing a significant challenge to antibiotic effectiveness and public health. The rise of carbapenem resistance is attributed to extensive and often unnecessary antibiotic use, particularly in developing countries, leading to limited antibiotic options. Carbapenem resistance can originate from 2 main mechanisms: β-lactam ring hydrolysis facilitated by carbapenemase enzymes or alterations in membrane permeability due to mutations in efflux pumps or porins, often in conjunction with extended-spectrum β-lactamase expression in Enterobacteriaceae [4].The Ambler classification system categorizes carbapenemases into 3 main classes: class A, class B (metallo-β-lactamases), and class D [5,6]. Class A carbapenemase, known as KPC1 (Klebsiella producing carbapenemase), is a type of enzyme that plays a significant role in conferring resistance to carbapenem antibiotics. Class B metallo-β-lactamases (MBLs), including New Delhi MBL (NDM), Verona integron encoded MBL (VIM) and Imipenemase (IMP), are enzymes that render antibiotics ineffective by hydrolyzing them. Class D Carbapenemases, also known as OXA-type enzymes, are characterized by their ability to hydrolyze carbapenems using a serine residue in their active sites [7]. Class D Carbapenemases play a crucial role in the global dissemination of carbapenem resistance, emphasizing the importance of surveillance, infection control measures, and the development of novel treatment strategies to address this urgent public health concern [8]. Carbapenem-resistant Enterobacteriaceae carrying the bla NDM gene are frequently identified in regions such as Asia, Africa, China, and the United Kingdom. On the other hand, the bla KPC gene has been have spread widely in the United States and South America [9]. In contrast, K. pneumoniae strains producing bla OXA48 are considered endemic to North Africa and the Middle East [8]. This distribution highlights the varied prevalence of different carbapenemase genes in different geographical regions, emphasizing the need for region specific strategies to monitor and address the spread of antibiotic resistance.Understanding the prevalence and distribution of carbapenem resistance genes is crucial for developing effective strategies to combat the antibiotic resistance.The aim of this study was to investigate the prevalence of carbapenem resistance in gram-negative bacteria isolated from patients admitted to Children's Medical Center Hospital. This study seeks to provide a comprehensive understanding of the molecular mechanisms contributing to carbapenem resistance.
- Methods
- Methods
- 1. Bacterial identification and antibiotic susceptibility testing
- 1. Bacterial identification and antibiotic susceptibility testing
Bacterial identification involved Gram staining for morphological assessment and differential biochemical tests. Organism identification utilized conventional biochemical methods, including oxidase, sugar fermentation, methyl red/Voges-Proskauer test, growth on Kliger’s Iron Agar, urease and a motility test [10-13].Antibiotic susceptibility testing was conducted using Escherichia coli ATCC 25922 as the quality control strain, and results were interpreted following the guidelines set by the Clinical and Laboratory Standards Institute [14]. In certain situations, the laboratory may have tested only a subset of antibiotics due to various factors such as resource constraints, clinical indications, or laboratory protocols. As a result, the number of isolates documented for each antibiotic may vary depending on the antibiotics included in the testing panel.- 2. Polymerase chain reaction amplification
- 2. Polymerase chain reaction amplification
Genomic DNA extraction from bacterial samples was conducted using a DNA extraction kit (Bioneer, Daejeon, Korea) following the manufacturer's instructions.The polymerase chain reaction (PCR) method was used for detection of the target genes including bla OXA23, bla OXA24, bla OXA48, bla OXA51, bla OXA58, bla OXA143, bla KPC, bla IMP, bla VIM, and bla NDM [6,15-20]. The PCR test was performed in a final volume of 25 μL with a final DNA concentration of 100 ng/mL. The PCR cycle consisted of an initial denaturation at 94°C for 5 minutes, followed by 35 cycles at 94°C for 30 seconds, annealing at 51°C–59°C for 40 seconds, and extension at 72°C for 40 seconds [13].- 3. Statistical analysis
- 3. Statistical analysis
The data were analyzed using IBM SPSS Statistics ver. 22.0 (IBM Co., Armonk, NY, USA). Categorical variables were presented as frequency and percentage.
The study was conducted at Children's Medical Center, a prominent and highly specialized Iranian hospital known for its expertise in pediatric care from June 2019 to June 2020. This hospital serves a significant patient population, attending to over 10,000 patients every month. Notably, Children’s Medical Center is recognized for delivering high-quality and specialized therapeutic services to neonates, infants, and children, not only within Iran but also across the broader region. The hospital currently operates a pediatric intensive care unit (ICU) with 10 beds and an emergency ICU established with 9 beds to enhance Emergency Department coverage. Additionally, the recently established infant ICU addresses the specific needs of acutely ill infants.During the period spanning from, a total of 777 gram-negative strains were isolated. The inclusion criteria for the selection of samples were as follows: Firstly, samples were considered if they exhibited a positive culture outcome, signifying the presence of microbial growth. Secondly, samples were included based on the presence of symptoms indicative of infection. We excluded duplicate strains from our study, choosing to incorporate only the initial strain when 2 or more strains were isolated concurrently. We did not differentiate between hospital-acquired and community-acquired cases in our enrollment criteria.Demographic information of patients, comprising details such as gender, age, length of hospital stays, and assigned wards, were collected.The need for ethical review and approval was waived because of the retrospective nature and anonymization of the analyzed data.
- Results
- Results
- 1. Antimicrobial resistance among carbapenem-resistant gram-negative bacteria
- 1. Antimicrobial resistance among carbapenem-resistant gram-negative bacteria
The overall analysis revealed an antibiotic resistance rate of 82.8% for sulfamethoxazole-trimethoprim, 43.9% for amikacin, 86.8% for cefepime, 90.8% for cefotaxime, 25.5% for ciprofloxacin, and 61.2% for piperacillin across the bacterial species evaluated. Notably, K. pneumoniae exhibited resistance in the following order: cefotaxime (93.8%), cefepime (87.5%), piperacillin-tazobactam (75.0%), sulfamethoxazole-trimethoprim (75.0%), amikacin (75.0%), and ciprofloxacin (37.5%). Similarly, E. coli demonstrated resistance to cefotaxime (90.1%), sulfamethoxazole-trimethoprim (87.7%), cefepime (86.4%), piperacillin-tazobactam (49.4%), amikacin (24.7%), and ciprofloxacin (11.1%) (Table 3).P. aeruginosa displayed resistance to ciprofloxacin (66.7%), amikacin (66.7%), and piperacillintazobactam (66.7%). Enterobacter spp. exhibited substantial resistance, with complete resistance to cefotaxime (100%), sulfamethoxazole-trimethoprim (100%), and piperacillintazobactam (100%). Salmonella spp. exhibited resistance to sulfamethoxazole-trimethoprim (100%). The highest antibiotic resistance rates were observed in A. baumannii, which displayed complete resistance to cefotaxime, cefepime, piperacillintazobactam, and amikacin.
During the 1-year study period, 141 carbapenem-resistant bacteria were isolated from a total of 777 bacterial isolates. E. coli was predominant, constituting 36.6% with 285 isolates, followed by K. pneumoniae (n=140, 18%), P. aeruginosa (n=112, 14.4%), Enterobacter spp. (n=50, 6.4%), A. baumannii (n=31, 4.0%), Serratia marcescens (n=32, 4.1%), and Salmonella spp. (n=24, 3.1%). Klebsiella spp., Citrobacter spp., Proteus mirabilis, Stenotrophomonas maltophilia, Pseudomonas spp., and Burkholderia spp. ranged from 0.9% 5.5%, while Morganella morganii represents a minimal 0.1%. Among carbapenem-resistant bacteria, E. coli stands out as the predominant contributor with 81 cases (57.4% of total), followed by K. pneumoniae with 16 isolates (15%) and A. baumannii with 15 isolates (10.6%). Other notable contributors include Salmonella spp. (3.5%), Enterobacter spp. (5.7%), and S. maltophilia (2.8%). Citrobacter spp., P. mirabilis, and P. aeruginosa have fewer isolates, while Pseudomonas spp., Burkholderia spp., and M. morgani show no carbapenem resistance (Fig. 1).The median age of the patients with carbapenem-resistant bacteria was 2 years (interquartile range, 3 months–5 years). The highest frequency was observed among children aged 1 year and under, representing 49.6 % of the total population (n=70), followed by children aged 1 to 5 years, accounting for 27% (n=38). Carbapenem-resistant bacteria were isolated from 77 boys (54.6%). They were cultured from various clinical specimens, including urine, Foley catheter, central venous catheter, surgery infusion, and bronchoalveolar lavage fluid. The distribution of isolates across these specimens revealed that the highest frequency was observed in urine samples (n=68, 48.2%) (Table 1). The distribution of bacterial isolates across different hospital wards revealed distinct patterns. The ICUs exhibited the highest prevalence at 27.7%, followed by the emergency ward at 21.3%, and the urology ward at 17.7% (Table 1). The remaining wards contribute to the distribution, each representing a distinct proportion ranging from 2.1% to 6.4%. K. pneumoniae emerges as a prominent bacterium in ICUs (50%) and surgery ward (25%). Meanwhile, E. coli was prevalent in urology (24.7%), and emergency ward (24.7%).The detailed distribution of carbapenem resistance genes among bacterial species is provided in Table 2 and Fig. 2. Notably, bla OXA48 showed the highest prevalence, being present in 33% of the total isolates, bla OXA143 and bla OXA58 also demonstrated substantial prevalence, accounting for 27% and 24%, respectively. bla OXA24 was present in 11% of the total isolates, bla OXA23 in 10%, and bla NDM in 10%. bla KPC, bla VIM, and bla IMP were not found in the analyzed bacterial isolates.Fig. 3 shows the diversity of carbapenem resistance genes and their combinations. In K. pneumoniae, prevalent carbapenemase genes include bla OXA58 (94%), bla OXA48 (31%), and bla NDM (19%). For E. coli, a diverse profile was observed, with significant occurrences of bla OXA143 (23%), bla OXA48 (41%), and bla NDM (7%).P. aeruginosa displayed varying percentages of carbapenemase genes, with bla OXA23 (33%) and bla OXA143 (67%) being prominent. Enterobacter spp. shows a mix of carbapenemase genes, including bla OXA58 (50%) and bla NDM (38%). A. baumanii demonstrates a high occurrence of bla OXA58 (73%).Klebsiella spp. presented high percentages of bla OXA143 (40%) and bla NDM (40%). P. mirabilis and S. marcescens each have isolates with carbapenemase genes, including bla OXA43 (100%) in P. mirabilis and bla OXA143 (100%) in S. marcescens. Citrobacter spp. showed carbapenemase genes, including bla OXA143 (100%).
- Discussion
- Discussion
The surge in antimicrobial resistance among gram-negative bacteria, especially in developing countries like Iran, poses a significant concern [17,21-28].In this study during a one-year period, it was observed that 18.1% of the gram-negative bacterial isolates exhibited phenotypic resistance to carbapenems. Notably, E. coli was the most prevalent bacterial strain, constituting 57.4% of the carbapenem-resistant cases. In comparison, K. pneumoniae and A. baumannii exhibited significantly lower frequencies, accounting for 11.3% and 10.6%, respectively. The prevalence of carbapenem resistance, especially in E. coli and A. baumannii, underscores the urgent need for effective infection control measures and the development of targeted therapeutic strategies to combat the rising threat of antibiotic resistance. The varied distribution among different bacterial species emphasizes the complex nature of resistance patterns and highlights the importance of continuous surveillance to monitor and address emerging trends in antibiotic resistance.Several research groups in Iran have previously published on carbapenem resistance rates and associated genes for specific gram-negative bacteria [21,29,30]. However, the available information remains limited. In our previous investigation focusing on carbapenem-resistant isolates exhibiting metallo-β-lactamase (MBL) production, 29% (32 isolates) of MBL-producing strains carried the bla NDM gene. Furthermore, the presence of the bla IMP and bla VIM genes was detected in 2% (2 isolates) and 1% (1 isolate) of MBL-producing strains, respectively [21]. Pourbaghi et al. [31], reported a lower resistance rate of 8% in Iran. Additionally, in a study conducted at a Moroccan hospital, it was found that enterobacteria comprised 52.7% of the bacterial isolates, with a corresponding ratio of 21.8% displaying resistance to carbapenems [32].Our study revealed a distinct prevalence of carbapenemase genes among the bacterial isolates. The diversity of carbapenem resistance genes and their combinations underscores the complexity of carbapenem resistance mechanisms. Notably, bla OXA48 showed the highest prevalence at 33%, followed by substantial occurrences of bla OXA143 (27%) and bla OXA58 (24%). The extensive presence of bla OXA48 across diverse species within hospitals, and notably in community settings, poses a significant challenge in constraining and managing the dissemination of bacteria carrying these enzymes. This difficulty arises from the capability of plasmids harboring bla OXA48 to readily and extensively transfer horizontally between different bacterial species [8]. This horizontal transmission mechanism contributes to the widespread distribution of the gene, making it a challenging factor for controlling the spread of carbapenem-resistant bacteria in various environments. Within K. pneumoniae, the predominant carbapenemase genes were identified as bla OXA58 (94%), bla OXA48 (31%), and bla NDM (19%). Klebsiella spp. demonstrated substantial proportions of bla OXA143 (40%) and bla NDM (40%). Interestingly, these findings differ from those reported by Jafari et al., where in K. pneumoniae, bla OXA48 took precedence as the most frequent carbapenemase gene at 72%, followed by bla NDM at 31% [30]. These variations underscore the complicated dynamics of carbapenem resistance, influenced by geographic. However, further investigation is needed to bridge this gap and establish comprehensive insights into the evolving patterns of carbapenem resistance, emphasizing the necessity for ongoing and region-specific surveillance initiatives to inform effective strategies for mitigating this global health concern.This development of carbapenem-resistant strains not only restricts the effectiveness of primary β-lactam antibiotics but extends its impact to other critical therapeutic molecules, including sulfamethoxazole-trimethoprim, aminoglycosides, and fluoroquinolones [33].The elevated rates of antibiotic resistance observed among carbapenem-resistant gram-negative bacilli present a challenge in the effective management of infections associated with these bacteria. The comprehensive analysis reveals alarming resistance rates, notably 82.8% for sulfamethoxazole-trimethoprim, 43.9% for amikacin, 86.8% for cefepime, 90.8% for cefotaxime, 25.5% for ciprofloxacin, and 61.2% for piperacillin-tazobactam across the diverse bacterial species. For E. coli, ciprofloxacin presents the lowest resistance at 11.1%, suggesting it may be a more favorable choice, followed by amikacin (24.7%). In the case of K. pneumoniae, considering the lower resistance rates, ciprofloxacin (37.5%) emerges as a more effective option.Our study has several limitations. Our study did not investigate the clinical outcomes or the impact of carbapenemase-producing isolates on patient morbidity and mortality, which would provide a more comprehensive perspective on the clinical implications of these resistant strains. Furthermore, the lack of information on patient specific factors, such as comorbidities and recent antibiotic use, hinders a more comprehensive analysis of risk factors associated with carbapenem resistance. We did not differentiate between hospital-acquired and community-acquired cases. Furthermore, it is imperative to acknowledge that our study is single-institutional and lacks prospective surveillance data.In conclusion, our study highlights the concerning prevalence of carbapenemase-producing gram-negative isolates among patients at the Children's Medical Center hospital. The notable resistance patterns observed, especially in K. pneumoniae and E. coli, underline the urgent need for proactive interventions, including appropriate antibiotic prescribing practices and the strengthening of antibiotic stewardship programs.
- Footnotes
-
Conflicts of interest The authors declare no conflict of interest.
Funding This study was supported by a grant (grant number: 1400314956626) from the Tehran University of Medical Sciences to Dr. Shima Mahmoudi. SM2 receiv ed partial support from the European CommissionEuro pean Research Executive Agency (REA) under Grant Agreement No. 101130873.
Author contribution Conceptualization: SM, SM; Data curation: SM, BP; Formal analysis: SM, RHS; Funding acquisition: SM; Methodology: SM, SM, BP, SP, RHS, Sadaf SM, MSA; Project administration: SM, SM; Visualization: SM, RHS; Writing original draft: SSM, SM; Writing review & editing: SM, SM, BP, RHS, SP, SSM, MSA
-
Fig. 1.

Fig. 2.

Fig. 3.
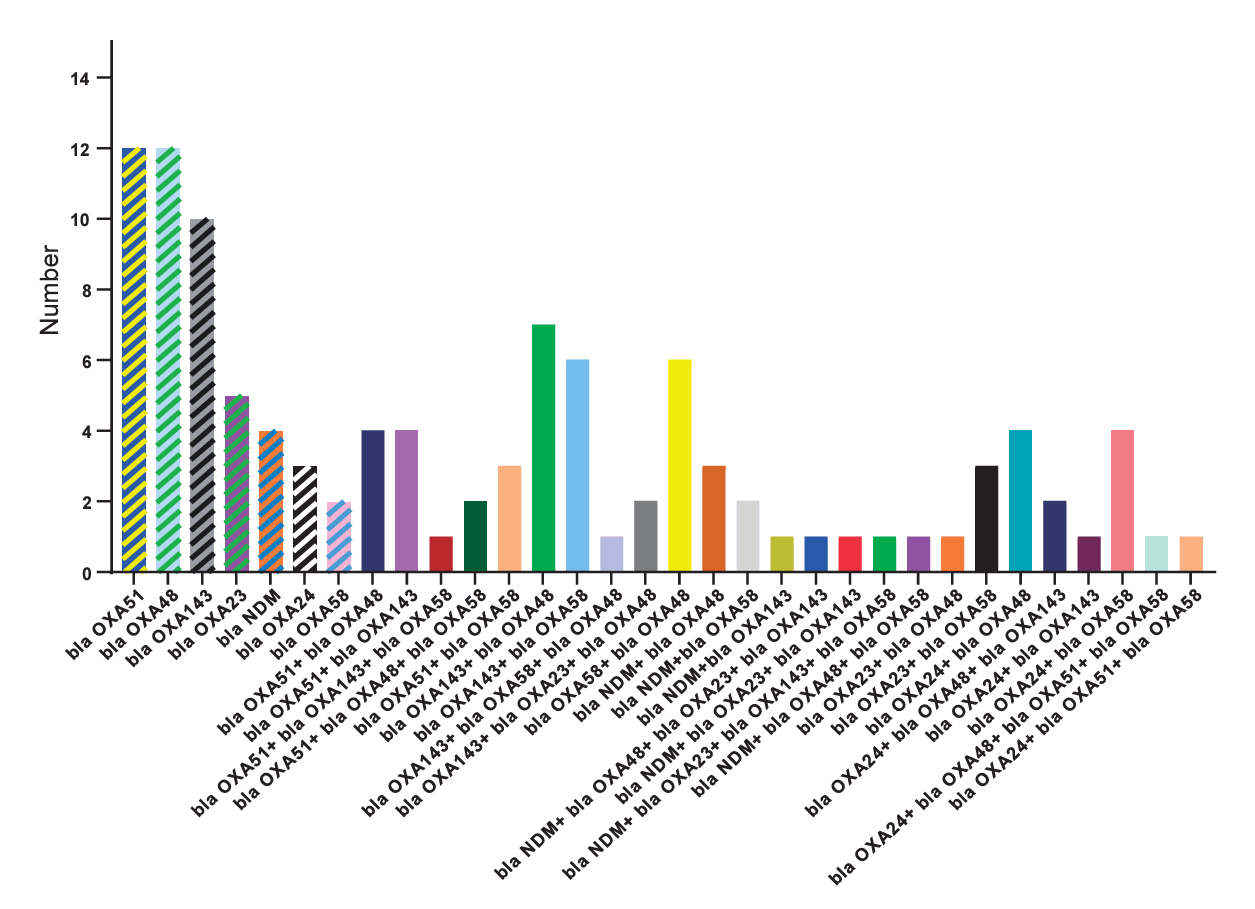
Table 1.
Table 2.
Table 3.
- References
- 1. Zeng M, Xia J, Zong Z, Shi Y, Ni Y, Hu F, et al. Guidelines for the diagnosis, treatment, prevention and control of infections caused by carbapenem-resistant gram-negative bacilli. J Microbiol Immunol Infect 2023;56:653–71.
[Article] [PubMed]2. Potter RF, D’Souza AW, Dantas G. The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug Resist Updat 2016;29:30–46.
[Article] [PubMed] [PMC]3. Salehi B, Abu-Darwish M, Tarawneh A, Cabral C, Gadetskaya A, Salgueiro L, et al. Antimicrobial resistance collaborators global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 2022;399:629–55.
[PubMed] [PMC]4. Logan LK, Weinstein RA. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis 2017;215(suppl_1): S28–36.
[Article] [PubMed] [PMC]5. Bush K. Classification for β-lactamases: historical perspectives. Expert Rev Anti Infect Ther 2023;21:513–22.
[Article] [PubMed]6. Yoshioka N, Hagiya H, Deguchi M, Hamaguchi S, Kagita M, Nishi I, et al. Multiplex real-time pcr assay for six major carbapenemase genes. Pathogens 2021;10:276.
[Article] [PubMed] [PMC]7. Kedišaletše M, Phumuzile D, Angela D, Andrew W, Mae NF. Epidemiology, risk factors, and clinical outcomes of carbapenem-resistant Enterobacterales in Africa: a systematic review. J Glob Antimicrob Resist 2023;35:297–306.
[Article] [PubMed]8. Pitout JDD, Peirano G, Kock MM, Strydom KA, Matsumura Y. The global ascendency of OXA-48-type carbapenemases. Clin Microbiol Rev 2019;33:e00102–19.
[Article] [PubMed] [PMC]9. Cui X, Zhang H, Du H. Carbapenemases in Enterobacteriaceae: detection and antimicrobial therapy. Front Microbiol 2019;10:1823.
[Article] [PubMed] [PMC]10. Jalalvand K, Shayanfar N, Shahcheraghi F, Amini E, Mohammadpour M, Babaheidarian P. Evaluation of phenotypic and genotypic characteristics of carbapnemases-producing enterobacteriaceae and its prevalence in a referral hospital in Tehran city. Iran J Pathol 2020;15:86–95.
[Article] [PubMed] [PMC]11. Kashkouri N, Tabarsi P, Pourabdollah Toutkaboni M, Kazempour Dizaji M, Bahrami N, Narimani A, et al. The prevalence of carbapenemase genes in carbapenem-resistant gram-negative bacilli, Masih Daneshvari Hospital, Tehran, Iran, 20192020. Iran J Med Microbiol 2022;16:573–80.
[Article]12. Chong Y, Shimoda S, Shimono N. Current epidemiology, genetic evolution and clinical impact of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect Genet Evol 2018;61:185–8.
[Article] [PubMed]13. Hosseinzadeh Z, Ebrahim-Saraie HS, Sarvari J, Mardaneh J, Dehghani B, RokniHosseini SMH, et al. Emerge of bla NDM-1 and bla OXA-48-like harboring carbapenem-resistant Klebsiella pneumoniae isolates from hospitalized patients in southwestern Iran. J Chin Med Assoc 2018;81:536–40.
[Article] [PubMed]14. Clinical and Laboratory Standards Institute. The Clinical and Laboratory Standards Institute performance standards for antimicrobial susceptibility testing; M100-ED30. Malvern (PA): Clinical and Laboratory Standards Institute, 2020.15. Hashemizadeh Z, Hatam G, Fathi J, Aminazadeh F, Hosseini-Nave H, Hadadi M, et al. The spread of insertion sequences element and transposons in carbapenem resistant Acinetobacter baumannii in a hospital setting in Southwestern Iran. Infect Chemother 2022;54:275–86.
[Article] [PubMed] [PMC]16. Hou C, Yang F. Drug-resistant gene of blaOXA-23, blaOXA-24, blaOXA-51 and blaOXA-58 in Acinetobacter baumannii. Int J Clin Exp Med 2015;8:13859–63.
[PubMed] [PMC]17. Mahmoudi S, Pourakbari B, Hosseini M, Alyari AE, Ashtiani MTH, Mamishi S. Molecular analysis of Pseudomonas aeruginosa metallo-beta-lactamase: a first report of an Iranian Referral Pediatric Hospital. Infect Disord Drug Targets 2018;18:46–51.
[Article] [PubMed]18. Wang TH, Leu YS, Wang NY, Liu CP, Yan TR. Prevalence of different carbapenemase genes among carbapenem-resistant Acinetobacter baumannii blood isolates in Taiwan. Antimicrob Resist Infect Control 2018;7:123.
[Article] [PubMed] [PMC]19. Grover SS, Doda A, Gupta N, Gandhoke I, Batra J, Hans C, et al. New Delhi metallo-β-lactamase - type carbapenemases producing Escherichia coli isolates from hospitalized patients: a pilot study. Indian J Med Res 2017;146:105–10.
[PubMed] [PMC]20. Pourgholi L, Farhadinia H, Hosseindokht M, Ziaee S, Nosrati R, Nosrati M, et al. Analysis of carbapenemases genes of carbapenem-resistant Klebsiella pneumoniae isolated from Tehran heart center. Iran J Microbiol 2022;14:38–46.
[Article] [PubMed] [PMC]21. Mahmoudi S, Pourakbari B, Rostamyan M, Raji H, Sadeghi RH, Mamishi S. Antimicrobial resistance patterns of gram-negative bacteria in an Iranian Referral Pediatric Hospital: a present danger of New Delhi metallo-β-lactamase. Infect Disord Drug Targets 2023;23:e180423215994.
[Article] [PubMed]22. Afsharipour M, Mahmoudi S, Raji H, Pourakbari B, Mamishi S. Three-year evaluation of the nosocomial infections in pediatrics: bacterial and fungal profile and antimicrobial resistance pattern. Ann Clin Microbiol Antimicrob 2022;21:6.
[Article] [PubMed] [PMC]23. Mahmoudi S, Mahzari M, Banar M, Pourakbari B, Haghi Ashtiani MT, Mohammadi M, et al. Antimicrobial resistance patterns of Gram-negative bacteria isolated from bloodstream infections in an Iranian referral paediatric hospital: a 5.5-year study. J Glob Antimicrob Resist 2017;11:17–22.
[Article] [PubMed]24. Mahmoudi S, Pourakbari B, Moradzadeh M, Eshaghi H, Ramezani A, Haghi Ashtiani MT, et al. Prevalence and antimicrobial susceptibility of Salmonella and Shigella spp. among children with gastroenteritis in an Iranian referral hospital. Microb Pathog 2017;109:45–8.
[Article] [PubMed]25. Mahmoudi S, Pourakbari B, Rahbarimanesh A, Abdosalehi MR, Ghadiri K, Mamishi S. An outbreak of ESBL-producing Klebsiella pneumoniae in an Iranian Referral Hospital: epidemiology and molecular typing. Infect Disord Drug Targets 2019;19:46–54.
[Article] [PubMed]26. Mamishi S, Mahmoudi S, Naserzadeh N, Hosseinpour Sadeghi R, Haghi Ashtiani MT, Bahador A, et al. Antibiotic resistance and genotyping of gram-negative bacteria causing hospital-acquired infection in patients referred to Children's Medical Center. Infect Drug Resist 2019;12:3377–84.
[PubMed] [PMC]27. Mamishi S, Shalchi Z, Mahmoudi S, Hosseinpour Sadeghi R, Haghi Ashtiani MT, Pourakbari B. Antimicrobial resistance and genotyping of bacteria isolated from urinary tract infection in children in an Iranian Referral Hospital. Infect Drug Resist 2020;13:3317–23.
[PubMed] [PMC]28. Pourakbari B, Mahmoudi S, Habibi R, Ashtiani MTH, Sadeghi RH, Khodabandeh M, et al. An increasing threat in an Iranian Referral Children's Hospital: multidrug-resistant Acinetobacter baumannii. Infect Disord Drug Targets 2018;18:129–35.
[Article] [PubMed]29. Khorvash F, Yazdani MR, Soudi AA, Shabani S, Tavahen N. Prevalence of acquired carbapenemase genes in Klebsiella pneumoniae by multiplex PCR in Isfahan. Adv Biomed Res 2017;6:41.
[Article] [PubMed] [PMC]30. Jafari Z, Harati AA, Haeili M, Kardan-Yamchi J, Jafari S, Jabalameli F, et al. Molecular epidemiology and drug resistance pattern of carbapenem-resistant Klebsiella pneumoniae isolates from Iran. Microb Drug Resist 2019;25:336–43.
[Article] [PubMed]31. Pourbaghi E, Doust RH, Rahbar M, Farzami MR. Investigation of OXA-23, OXA-24, OXA-40, OXA-51, and OXA-58 genes in carbapenem-resistant Escherichia coli and Klebsiella pneumoniae isolates from patients with urinary tract infections. Jundishapur J Microbiol 2022;15:e119480.
[Article]

 About
About Browse articles
Browse articles For contributors
For contributors

