Next-generation sequencing (NGS) technologies are massively parallel DNA sequencing methods that allow faster and cheaper sequencing of nucleotides than does Sanger sequencing [1]. With the advent of NGS, nucleotide sequencing has found applications in the diagnosis of pediatric neurological disorders such as muscle disease, epilepsy, neurodegenerative disease, neurometabolic disease, and autism spectrum disorder [2-4]. In recent years, several studies on the genetic diagnosis of unexplained global developmental delay (DD) and intellectual disability (ID) have been published [5-7]. Global DD and ID have a high prevalence, affecting 1%–3% of children [8]. The diversity of phenotypical and genetical heterogeneity leads to a low diagnostic yield and insufficient clinical management [7].
When the clinical picture suggests a syndromic form of DD/ID, a diagnostic genetic test is used to confirm or rule out chromosomal abnormalities or monogenic disorders: classical karyotyping or fluorescence in situ hybridization or molecular biology testing (FMR1 for fragile X syndrome, MECP2 for Rett syndrome) [2]. As high-resolution chromosomal microarray (CMA) became widely used and provided a much higher diagnostic yield than traditional karyotyping, it became the first-tier of genomic analysis for patients presenting with DD/ID in the 2010 American College of Medical Genetics guideline [9]. NGS technologies have recently become a key diagnostic tool for establishing a genetic diagnosis in children with DD/ID, reducing the mean cost per diagnosis in cases that are generally more challenging to detect, such as mosaicism [7].
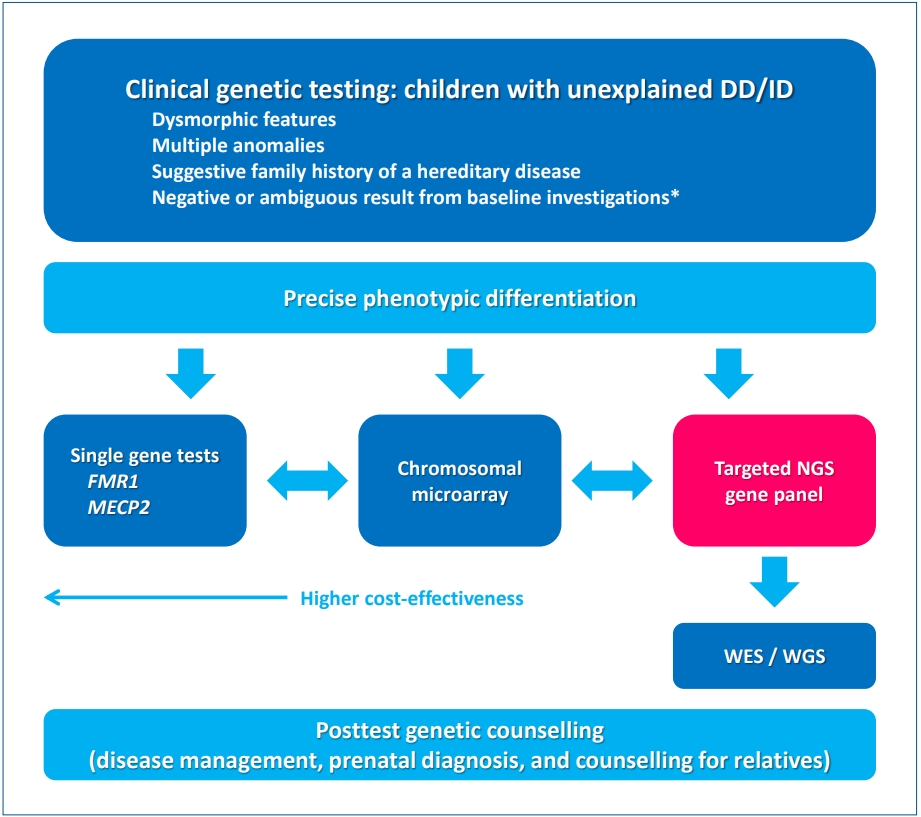
Han et al. [6] previously reported a diagnostic yield of 29% (10 of 25) in Korean patients with DD and/or ID. They suggested that targeted NGS through singleton analysis with a phenotypefirst approach is useful for genetic diagnosis. In this review article, Han emphasized a broad acceptance of NGS applications such as targeted panel sequencing (TPS), whole-exome sequencing (WES), and whole-genome sequencing (WGS) with reduced cost. Suggested guidelines for genetic investigations in children with DD/ID recommend TPS as the first-tier test and familybased trio testing by WES/WGS as the second-tier test, although a few technical limitations such as detecting microdeletions or duplications and mutations in noncoding trinucleotide expansion diseases [10]. Accordingly, an updated clinical approach to genetic investigations in children with DD/ID is shown in Fig. 1 [9,11]. The first step in increasing the diagnostic yield is to clearly define the population of suspected genetic causes that have dysmorphic features, multiple anomalies, suggestive family history of a hereditary disease, and negative or ambiguous results from baseline investigations. Another important consideration is that more detailed and precise phenotypic differentiations lead to a higher diagnostic yield of genetic diagnosis. After differentiating phenotypes, clinicians can choose the first-tier test with consideration of cost-effectiveness: single gene tests, CMA, or TPS. WES/WGS can help to identify a causative genomic variant in cases in which previous testing was inconclusive. Once a genetic diagnosis is established, posttest genetic counseling should address possible therapeutic interventions and the risk of recurrence.
There are still many considerations related to advanced genetic testing that continuously reclassifying variants of unknown significance and ethical issues [11]. Nonetheless, the role of NGS testing in the clinical setting is becoming more important and clinicians should be aware of its rapid changes in this genetic era.





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link PubMed
PubMed Download Citation
Download Citation