Article Contents
| Korean J Pediatr > Volume 55(10); 2012 |
Abstract
Purpose
Single-nucleotide polymorphism (SNP) markers within LIN28B have been reported to be related to the timing of pubertal growth. However, no study has investigated the frequency of genetic markers in girls with precocious puberty (PP) or early puberty (EP). This study aimed to determine the frequency of putative genetic markers in girls with PP or EP.
Methods
Genomic DNAs were obtained from 77 and 109 girls that fulfilled the criteria for PP and EP, respectively. The controls in this study were 144 healthy volunteers between 20 and 30 years of age. The haplotypes were reconstructed using 11 SNPs of LIN28B, and haplotype association analysis was performed. The haplotype frequencies were compared. Differences in the clinical and laboratory parameters were analyzed according to the haplotype dosage.
Results
Eleven SNPs in LIN28B were all located in a block that was in linkage disequilibrium. The haplotype could be reconstructed using 2 representative SNPs, rs4946651 and rs369065. The AC haplotype was less frequently observed in the PP group than in the controls (0.069 vs. 0.144, P=0.010). The trend that girls with non-AC haplotypes tended to have earlier puberty onset (P=0.037) was illustrated even in the EP+PP patient group by Kaplan-Meier analysis.
Central precocious puberty (PP) is characterized by premature activation of the hypothalamic GnRH pulse generator with subsequent pulsatile gonadotropin secretion. In general, PP is defined at the lower age limit of eight years of age in girls (presenting with Tanner stage B2) and nine years of age in boys (Tanner G2 and/or testicular volume 4 mL)1-3). Children with pubertal development just above these age limits (girls with B2 between 8 and 9 years of age) are considered to have early puberty (EP); however, some of these children have similar concerns to children with PP.
Genetic predisposition is considered to affect PP and EP in girls. Recently, several genome wide association analyses identified common genetic markers related to menarche near the LIN28B gene at 6q214-7). Ong et al.4) reported that rs314276 or another related variant within LIN28B could be significant genetic markers associated with the timing of pubertal growth and development both in girls and boys. LIN28B is known as a potent and specific regulator of microRNA (miRNA) processing controlling the timing of developmental events8).
In the present study, the trend of genetic association of the timing of puberty observed in general population was thought to also be observed or more exaggerated in girls with PP or EP. Therefore, the genetic association of girls with EP and PP, using single nucleotide polymorphism (SNP) markers in the LIN28B gene, was studied.
From January 2010 to July 2010, girls referred to the Pediatric endocrinology clinic at this center for early pubertal development were enrolled. Pubertal and sexual development were evaluated according to Tanner stages. PP was diagnosed in girls with breast development before eight years of age, advanced bone age, and a pubertal response on the GnRH test. Advanced bone age was defined as an increase in the bone age by more than 1 year compared to the chronological age. A pubertal response on the GnRH test was defined as a peak LH over 5 mIU/mL and a peak LH level 2 to 3 times more than the basal level. EP was defined in girls with more than a Tanner stage 2 breast development between 8 and 9 years of age, advanced bone age and a pubertal response on the GnRH test. A total of 77 girls and 109 girls fulfilled the criteria for PP and EP, respectively. The clinical and patient-related data for the girls with EP and PP are presented in Table 1.
The controls in this study were 144 healthy volunteers between 20 and 30 years of age; their medical examination records were available. Their mean age at menarche (12.7┬▒1.2 years), adult height (161.7┬▒4.1 cm) and weight (51.7┬▒4.3 kg) did not significantly deviate from the Korean population standards. This study was reviewed and approved by the Institutional Review Board (IRB File No.: 2009-12-070-001) and informed consents were obtained from the subjects and/or their parents.
Whole blood specimens were collected from each individual using ethylenediaminetetraacetic acid tubes and genomic DNA was isolated from peripheral blood leukocytes using the Wizard Genomic DNA Purification kit according to the manufacturer's instructions (Promega, Madison, WI, USA). After a review of the medical literature, 11 significant SNPs (P<5├Ќ10-8) around LIN28B were selected. Multiplex SNP genotyping was performed by primer extension and matrix-assisted laser desorption/ionization time-of-flight, and mass spectrometry using iPLEX Gold technology from Sequenom (Sequenom Inc., San Diego, CA, USA). SNP assays were designed using Sequenom's Real SNP (www.RealSNP.com) and MassARRAY Assay Design ver. 3.0 (primer information is available upon request). Polymerase chain reaction was performed using 20 ng of DNA in a 10 mL reaction volume for 35 cycles with standard iPLEX methodology. Spectra were analyzed using the MassARRAYTyper ver. 3.4 (Sequenom Inc.). Excluding individual SNPs or samples with call rates by genotype below 95% was used for quality control and SNP assays with poor quality spectra/cluster plots.
The clinical characteristics of the study participants are expressed as means┬▒standard deviations (SD). SNPs with Hardy-Weinberg equilibrium and P values greater than 0.05 were regarded as significant. Haplotype and statistical analyses were performed using the SNP and Variation Suite (SVS) 7 (Golden Helix Inc., Bozeman, MA, USA). Haplotype reconstruction and frequency estimation were carried out using the composite haplotype method (CHM) algorithm. A logistic regression analysis was performed to compare the CHM frequency differences between cases and controls. A regression analysis was performed to compare the differences in clinical and laboratory parameters among different haplotype dosages. A Kaplan-Meier analysis was used to identify differences in pubertal onset according to haplotype dosages. Correction for multiple testing was carried out using the full-scan permutation method with 1,000 replicates. A P value of <0.05 was considered statistically significant.
Clinical and patient-related parameters including age at diagnosis, bone age at diagnosis, height, weight and body mass index (BMI) standard deviation scores (SDSs) in the PP and EP groups are presented in Table 1. There were positive deviations in height, weight and BMI SDSs in both the PP and EP groups, reflecting the effect of sex hormone exposure on body mass. However, no significant differences were between the PP and EP groups. Their hormone levels including E2, follicle-stimulating hormone and luteinizing hormone approximated the adolescent range; however, there was no significant difference between the two groups.
Both in cases and controls, 10 consecutive SNPs from the 5'-most SNP, rs9391253, were in very high linkage disequilibrium (LD), and all the 11 SNPs analyzed were located within one LD block (Fig. 1). Therefore, the haplotypes were reconstructed and three main haplotypes were found that could be represented by two SNPs, rs4946651 and rs369065. Haplotype association analysis revealed that the AC haplotype was less frequently observed in the PP group compared to the controls (0.069 vs. 0.144, P=0.010; corrected P=0.039), suggesting that AC haplotype might be associated with later pubertal onset. However, such trend was not noted in the EP group (Table 2).
The most probable haplotypes were reconstructed in each individual and they were classified according to the AC haplotype dosage. A regression analysis showed that patients with non-AC haplotypes were associated with a lower age at diagnosis, with borderline significance (P=0.059); however, other clinical and laboratory parameters were not significantly correlated with the AC haplotype dosage (data not shown). The Kaplan-Meier analysis showed that AC haplotypes are associated with later pubertal onset (PP+EP; P=0.037) (Fig. 2).
Recently, variation in or near the LIN28B gene has been found associated with age at menarche in four independent genome-wide association (GWA) studies, and, in one of them, the allele associated with earlier age at menarche was also associated with other markers of pubertal timing (earlier breast development; earlier voice breaking and more advanced pubic hair development; faster tempo of height growth and shorter adult height)4-7). A second menarche locus was identified in an intergenic region at 9q31.28,10. These two loci together explained only 0.6% of the variance in age at menarche4-7). And recent GWA study identified new thirty loci for age at menarche. In total, 945 SNPs representing 45 loci were associated with age at menarche at genome-wide significance levels. But the two most significant loci for age at menarche confirmed the previously reported associations at LIN28B (rs775993)8) and 9q31.2 (rs2090409)9).
Otherwise, rs314280 and rs2090409 polymorphisms are not a useful marker for prediction of the susceptibility of PP in Taiwanese girls10). This result is not consistent with the previous study4-7). The discrepancy may be due to different illness classifications, and racial and disease variations.
LIN28B is a human ortholog of the gene that regulates processing of miRNAs which control the timing of major developmental events in C. elegans: gain-of-function and loss-of-function mutations result in retarded or precocious development, respectively11). Thus, LIN28B may have a role in pubertal development and are good candidate genes for early pubertal development.
This is the first clinical study investigating the association of genetic markers with PP and EP in girls in Korea. Although the present study may have low statistical power (0.28 for ╬▒=0.05) due to the small number of samples, the results of this study showed that SNP markers in the LIN28B gene (6q21) were significantly associated with pubertal onset in girls with PP. Haplotype analysis revealed that SNPs in the 5' region to intron 2 of the LIN28B gene were in a strong LD block in the Korean cohort, similar to the findings in Caucasians and Asians from the International Hapmap Project12). The AC haplotype in this region was consistently associated with delayed pubertal onset, and the direction of the association was consistent with the finding of prior studies on the timing of menarche. Therefore, genetic variations in the LD block spanning the 5' part to intron 2 of LIN28B may be associated with the timing of puberty, not only in general population but also in patients with PP and EP. The trend was also observed even within the patient group; Kaplan-Meier analysis showed approximately 5 to 8 months delay of pubertal onset with LIN28B allele dosage dependence, which may be important with regard to the timing of exposure to female sex hormones.
Physical variables including height, weight and BMI are known to be related to the onset of puberty. Epidemiological studies have indicated that early menarche or puberty is associated with decreased final height, and notably, LIN28B is the only gene, among the many height-related genes, shown to be associated with the timing of puberty. Early exposure to sex hormones is thought to contribute to the final adult height rather than the height of the child predisposing to PP or EP. Similarly, the data in this study showed no height SDS difference according to the haplotype dosage. Weight-gain is known to affect menarche, with heavier girls maturing earlier. The results of this study showed no weight SDS difference according to the haplotypes. Instead, height, weight, and BMI SDS values in all subgroups tended to be higher than those of the normal population, which reflects the fact that all of the girls had entered puberty already.
In conclusion, the present study showed that haplotypes of LIN28B were significantly associated with PP in girls. The findings of this study add to the knowledge on the genetic aspects of the onset of puberty. Further studies are needed in other ethnic groups as well as additional basic research on the biology of the LIN28B gene.
Acknowledgment
This research was supported by a grant from the Samsung Genomic Center Research Fund (D-B0-007-1).
References
1. Marshall WA, Tanner JM. Variations in pattern of pubertal changes in girls. Arch Dis Child 1969;44:291РђЊ303.



2. Marshall WA, Tanner JM. Variations in the pattern of pubertal changes in boys. Arch Dis Child 1970;45:13РђЊ23.



3. Carel JC, Lahlou N, Roger M, Chaussain JL. Precocious puberty and statural growth. Hum Reprod Update 2004;10:135РђЊ147.


4. Ong KK, Elks CE, Li S, Zhao JH, Luan J, Andersen LB, et al. Genetic variation in LIN28B is associated with the timing of puberty. Nat Genet 2009;41:729РђЊ733.



5. Sulem P, Gudbjartsson DF, Rafnar T, Holm H, Olafsdottir EJ, Olafsdottir GH, et al. Genome-wide association study identifies sequence variants on 6q21 associated with age at menarche. Nat Genet 2009;41:734РђЊ738.


6. He C, Kraft P, Chen C, Buring JE, Pare G, Hankinson SE, et al. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat Genet 2009;41:724РђЊ728.



7. Perry JR, Stolk L, Franceschini N, Lunetta KL, Zhai G, McArdle PF, et al. Meta-analysis of genome-wide association data identifies two loci influencing age at menarche. Nat Genet 2009;41:648РђЊ650.



8. Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science 2008;320:97РђЊ100.



9. Elks CE, Perry JR, Sulem P, Chasman DI, Franceschini N, He C, et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat Genet 2010;42:1077РђЊ1085.



10. Chou IC, Wang CH, Lin WD, Tsai CH, Tsai FJ. Association study in Taiwanese girls with precocious puberty. J Pediatr Endocrinol Metab 2011;24:103РђЊ104.

Fig.┬а1
Haplotype analysis result indicating that 11 single-nucleotide polymorphisms from the 5'-region to intron 2 were in a strong linkage disequilibrium block with high r2 scores.
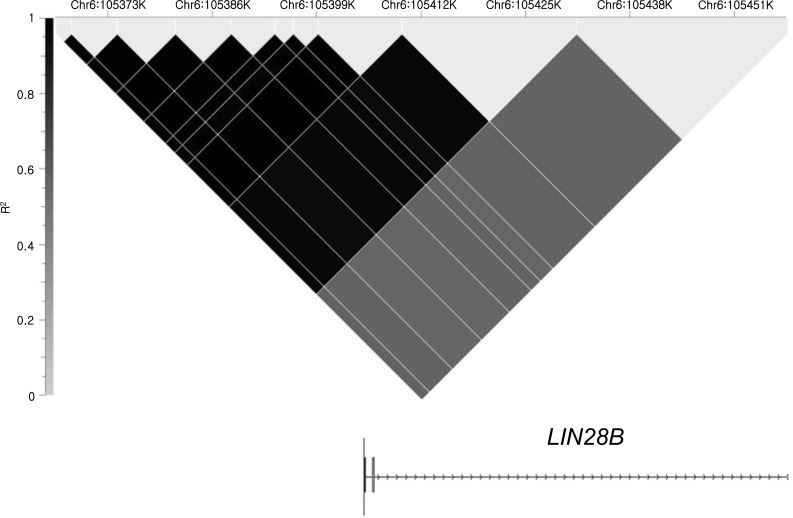
Fig.┬а2
Kaplan-Meier analysis results according to the AC haplotype dosages in the PP+EP patient group, illustrating that AC haplotype dosage (0, 1, and 2 alleles) is associated with late pubertal onset (P=0.037). PP, precious puberty (onset before 8 years of age); EP, early puberty (onset between 8 and 9 of ages).






 PDF Links
PDF Links PubReader
PubReader PubMed
PubMed Download Citation
Download Citation