Introduction
The human microbiome, which consists of a collective cluster of commensal, symbiotic, and pathogenic microorganisms living on the human body, plays a key role in host health and immunity [1]. The human nasal cavity harbors commensal bacteria that suppress the colonization of opportunistic pathogens. However, when a microbial community becomes dysfunctional or imbalanced, opportunistic pathogens in the bacterial microbiome of the nasopharyngeal or nasal sinus cavity can spread to adjacent regions of the respiratory tract and become involved in the development of diseases such as acute respiratory infections including otitis media (OM), sinusitis, and bronchitis and allergic respiratory diseases including asthma [2,3]. In fact, many environmental factors modulate the composition of the nasal microbiome [4,5], which is not constant throughout life; rather, it is dependent on the body’s habitat and host health status [6]. From this aspect, we review the factors influencing changes in the nasal microbiota, causalities with disease development, and immune alterations.
Nasal microbiota influencing factors and health
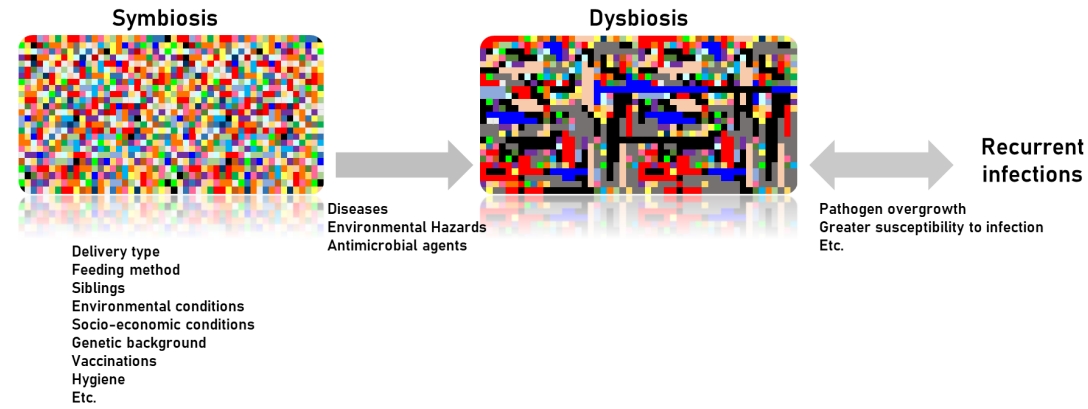
Many studies recently reported on the complexity of the nasopharyngeal microbiota in children and adults, which undergo significant changes upon exposure to disease, environmental hazards, and antimicrobial agents or vaccinations. In addition, delivery type, feeding or dietary habituation, smoking, lifestyle, pollution, and hygiene are well known influencing factors that modulate the nasal microbiota. These differences are related to genetic background variables and socioeconomic conditions such as housing, health care, poor hygiene, family size, overcrowded living conditions, day care contact, and number of siblings. In fact, newborns delivered by vaginal versus cesarean delivery are exposed to different microbiota [7], and early postnatal bacterial colonization of the upper airways is significantly affected by birth season [8]. A recent study reported that the nasal microbiota among dairy farmers is more complex than their oral microbiota, which protects against infections and competes with Staphylococcus aureus colonization (Table 1) [9].
The nasal microbiota consists of various microbial communities, including many different genera of aerobic and anaerobic microorganisms. Dysbiosis of the nasal microbiota may lead to pathogenic overgrowth and greater susceptibility to infection, as has been observed in the gastrointestinal tract and lower airways. In unhealthy situations, many respiratory pathogens, including resistant strains, may be reserved as nasopharyngeal microbiota and serve as a source for the transfer of antimicrobial-resistant genes from non-pathogenic to pathogenic bacteria [10]. Therefore, the abundance and diversity of nasal microbiota are prognostic factors of good health (Fig. 1) [7,11-13].
In conclusion, the factors influencing nasal colonization and elimination are not fully understood, but adhesion to nasopharyngeal mucosa receptors and immune responses are implicated in addition to the properties of the nasopharyngeal bacterial microbiota and the dynamics of colonization resistance. Overall, potential pathogens are more likely to colonize the nasopharynx of children with recurrent respiratory infections, including OM, due to their impaired local immunity and repeated exposure to respiratory pathogens as well as adults with chronic respiratory tract disease, who have higher carriage rates (Table 2) [14].
Nasopharyngeal microbiota with acute respiratory infections: focus on immune defense correlations
The early postnatal bacterial colonization of the upper airways is significantly affected by birth season, emphasizing a future need to focus on the seasonality aspect in models of the impact of early dynamic changes in airway bacterial communities in relation to later disease development. Nasopharyngeal colonization occurs early in life and is consistently present thereafter, although bacterial species and strains are transient, particularly with antibiotic use and pneumococcal vaccine routines [15-17].
The most important role of nasal mucosal immunity is to defend the adjacent respiratory organs against invading pathogens while continuing to maintain symbiotic relationships between extensive microbiota and the host [18]. The main bacterial nasopharyngeal microbiota of OM, Streptococcus pneumoniae, nontypeable Haemophilus influenzae, and Moraxella catarrhalis are most likely to engage in a virulence shift [19]. Such influences are multifactorial and include microbial interactions, microbial carriage load, mucosal integrity, demographic and environmental factors, and host immunity [20,21]. Moreover, lymphocyte expansion and maturation into effector lineages is influenced by antigen exposure and dose, whereby high-dose continual antigen exposure supports the differentiation and expansion of Treg lymphocytes [22-24].
One study suggested that circulating regulatory T lymphocytes may contribute to a host’s tolerance of nasopharyngeal colonization and the potential development of chronic OM [22]. From this aspect, it is very important to determine the relationships between these factors and host immune responses to gain an increased understanding of its pathogenesis.
The host immune response against invading respiratory viruses can provoke protective immunity in the respiratory microbiome, including the nasal microbiome [25]. An influenza A virus infection can modify the community structure of the respiratory microbiome by increasing the number of pathogenic bacteria. In fact, influenza A infections showed increased nasal carriage of S. pneumoniae and S. aureus in adults and showed nasal microbiota differences between healthy persons and influenza A-infected patients [26,27]. Other viruses such as adenovirus and rhinovirus can increase the colonization density of S. pneumoniae in the nasal and respiratory tract, continuously increasing the risk of pneumococcal infection [28]. Elevated H. influenzae or Moraxella nasal carriages were observed after several viral infections such as respiratory syncytial virus or rhinovirus in infants [29-31].
In conclusion, nasal microbiota dysbiosis is associated with respiratory viral pathogen invasion, escaping the host immune system and leading to respiratory diseases.
Nasopharyngeal microbiota with OM
Acute OM (AOM) is among the most common pediatric infectious diseases. The mechanism of bacterial AOM development is well known; a respiratory viral infection disrupts the mucociliary system, impairing the host’s primary mechanical defense against bacterial invasion, continuously leading to reduced middle ear pressure and forcing mucus, nasopharyngeal secretions, and bacteria colonies from the nasopharynx into the middle ear [31,32]. Although the disease is primarily considered a bacterial infection, it is also well known that viral upper respiratory tract infections (URIs) predispose children to AOM and viruses alone can cause AOM [33]. The nasopharyngeal bacterial microbiota in healthy individuals differs from nasopharyngeal colonization in individuals with URIs or AOM [34,35]. Children concurrently colonized with S. pneumoniae, nontypable H. influenzae, and M. catarrhalis are at higher risk of developing AOM than children without pathogenic bacteria. Children have more bacterial types and higher bacterial colony counts in the nasopharynx during URIs than during a healthy period. However, the majority of causative pathogens associated with AOM were newly acquired pathogens, not those found during the healthy period [35]. Finally, the effective prevention of AOM must include the prevention of viral URIs and the prevention and/or elimination of nasopharyngeal colonization with pathogenic bacteria [36]. However, to effectively prevent AOM, more studies are required to better understand how these interventions affect bacterial and viral interactions in the pathogenesis of viral AOM.
OM with effusion (OME) is the most common cause of hearing alterations in childhood [37]. S. pneumoniae, H. influenzae, and M. catarrhalis are the most common pathogens of OME based on classical culture approaches. With the recent advancement of culture-independent techniques such as 16S rRNA pyrosequencing, previously unknown bacterial communities have now been identified within the middle ear [38]. In fact, Alloiococcus, Haemophilus, Staphylococcus, Corynebacterium, Streptococcus, and Moraxella were proven as important pathogens in OME development using both traditional culture, culture-independent polymerase chain reaction, and 16S pyrosequencing techniques [38-41]. In addition, the adenoid microenvironment does not correlate with the middle ear microenvironment, and it has been confirmed that OME is not a sterile condition and casts doubt on the commonly held belief that the adenoid pad serves as a bacterial reservoir for OME, while the microenvironment of the middle ear itself may play a greater role in influencing the constitution of the microbial communities within them rather than bacteria from the nasopharynx. Moreover, differences between the local microbiota of the adenoid and the middle ear strongly suggest that the microenvironment of the middle ear plays a greater role in the composition of the microbiota than the potential bacterial seeding from the adenoid pad to the middle ear in children with OME [42]. However, further studies of the causal relationship between the microbiome of the middle ear and OME pathogenesis are needed (Table 2).
Nasopharyngeal microbiota with sinusitis
Acute and chronic rhinosinusitis are the most frequent complications in children with complicated diseases. Therefore, extensive studies have been performed. Among them, a number have clarified various microbiological aspects of rhinosinusitis, including its epidemiology and the role of bacterial biofilm producers. Studies of the microbiota of the nasosinus in healthy children and adults were recently conducted. However, no precise bacterial and/or viral etiology has yet been established and knowledge remains limited. A few studies suggested that microbiotal diversity increases significantly with age. Moreover, the ecological changes in the nasopharyngeal and nasal cavity sites may influence the development of adenoid hypertrophy or result from hypertrophy in healthy controls and patients with sinusitis [43,44].
Many researchers consider bacteria with or without biofilm “social organisms” that form the so-called microbiota. Sinonasal microbiota can modulate the course of both acute and chronic rhinosinusitis. As the composition, distribution, and abundance of microbiota in the nasosinus cavity impact mucosal health and can influence pathogenic growth and function, a greater understanding of the host–microbiome constituents and relationships may encourage the development of new treatments for acute or chronic rhinosinusitis [11]. The concentration, uniformity, and type/number of strains of sinonasal microbiota may vary among sites. The relative and total microorganism counts can also be affected by various factors [45,46]. In fact, the presence and percentage of microorganisms can be influenced by previous antimicrobial treatments, vaccinations, the presence of normal flora capable of interfering with pathogenic growth, geography, and site, as the maxillary, ethmoid, and frontal sinuses show different etiologies [47]. Therefore, it is difficult to use such data when deciding the choice of antibiotic treatment for controlling sinusitis.
The formation of biofilm in rhinosinusitis may play a significant role in its pathogenesis and persistence. Biofilm has a number of advantages in terms of bacterial survival, and their perpetuation can create a certain degree of instability in host–bacterial interactions. A number of aerobes and anaerobes are capable of producing biofilm, which differs depending on the microorganism and stimulates various inflammatory molecules during acute or chronic rhinosinusitis. The most negative features of biofilm are their high degree of resistance to antibiotics and host immune mechanisms, as they are less susceptible to opsonization and phagocytosis [48].
In conclusion, the relationship between microbiota and sinonasal diseases has been well characterized, but further studies are necessary to establish its implications for the bacterial network under pathological conditions.
Nasopharyngeal microbiota with pneumococcal infections
S. pneumoniae, also known as pneumococcus, is among the deadliest pathogens in humans and one of the most dominant causes of sepsis, pneumonia, OM, and sinusitis. Pneumococcus is colonized at some point in nearly all humans during childhood at various durations and frequencies by age [49]. However, most individuals do not develop pneumococcal diseases; rather, a perfect balance is maintained between the host, pneumococcus, and microbiota. The disruption of this balance alters the existence of pneumococcus as a commensal pathogen and is, therefore, called a pathobiont.
The nasopharyngeal acquisition of pneumococcus is the first step of virtually all pneumococcal diseases [50], while children with no bacteria in the nasopharynx are considered at low risk of developing disease [36]. The following 2 important questions arise: (1) can pneumococcal vaccines prevent colonization, which subsequently prevents pneumococcal diseases; and (2) which factors influence the change of pneumococcus from a simple colonizer to a virulent pathogen?
The concern of eliminating a population of bacteria from its niche would be replacing the void with other pathogens that could cause similar but more severe diseases [51]. There are currently 3 licensed pneumococcal vaccines in Korea: the 10-valent pneumococcal conjugate vaccine (PCV10), the 13-valent pneumococcal conjugate vaccine (PCV13), and the 23-valent pneumococcal polysaccharide vaccine (PPSV23). After the introduction of these vaccines, there have been many studies on the nasopharyngeal carriage rate, serotypes of S. pneumoniae, and antimicrobial susceptibilities. These studies show that although conjugate vaccines have been immensely successful at reducing and eliminating diseases caused by vaccine serotypes, the colonization rate has not changed; rather, there has been a replacement in the colonization of nasopharyngeal pneumococcal serotypes into nonvaccine types that show nonsusceptibility to penicillin and erythromycin [51-55].
The cocolonization and polymicrobial interaction of pneumococcus with other respiratory colonizers, mainly non-typeable H. influenzae and M. catarrhalis cause different risks regarding the development of OM [36]. In fact, in children who have concomitant nontypeable H. influenzae and S. pneumoniae in the nasopharynx, many pneumococcal serotypes demonstrate lower rates of progression to OM, and nontypeable H. influenzae is likely the cause in up to 75% of cases [56,57]. An animal model suggested that increased neutrophil recruitment and promotion of pneumococcal clearance, which then diminishes the ability of pneumococcus to progress to disease, is the primary mechanism [58]. Regarding the cocolonization by S. pneumoniae and S. aureus, no negative interaction was observed [26].
The coinfection with some respiratory viruses increases the rate of colonization with pneumococcus and causes high pneumococcal colonization densities, consequently having significant associations with invasive pneumococcal pneumonia [59]. There are also many studies of the synergistic effect of concomitant pneumococcal-influenza infections. Influenza virusinduced polymorphonuclear neutrophil dysfunction is a key component of influenza virus-induced secondary pneumococcal diseases; although the role of polymorphonuclear neutrophils is unclear, the data indicate that neutrophils may play both protective and damaging roles in response to pneumococcal infections (Table 2) [60].
Serotype is an important factor that determines the progression from pneumococcal colonization to disease, and some serotypes have a higher potential to cause invasive disease than others. However, unlike those causing invasive diseases, little difference was noted among serotypes in their ability to ascend from the nasopharynx to the middle ear and subsequently cause mucosal diseases [61,62], although there can be a difference of up to 100-fold inthe degree [63]. Altogether, this explains how conjugate vaccines do not reduce the prevalence of pneumococcal nasal carriage but decrease the incidence of invasive pneumococcal diseases and impact the incidence of acute and complex OM.
In conclusion, the current vaccines effectively reduce pneumococcal diseases caused by vaccine serotypes, especially invasive diseases. However, nonvaccine serotypes are replacing the niche, and further studies are needed to determine the overall burden of replaced serotypes on pneumococcal diseases, especially in mucosal diseases such as AOM. To the best of our knowledge, cocolonization and polymicrobial interaction of pneumococcus with other respiratory colonizers, coinfection with respiratory viruses, and serotypes are some factors that determine whether pneumococci remains colonizers or become pathogens.
Nasopharyngeal microbiota with allergic rhinitis and asthma
Many studies have suggested that the nasal microbiome may play an important role in the modulation of localized immune responses and development of respiratory allergic diseases such as allergic rhinitis and asthma [3,64-66]. In fact, the nasal microbiota before and during pollen seasons show a significant increase in bacterial biodiversity in the middle meatus in patients with seasonal allergic rhinitis compared to healthy controls and a correlation between an increase in symptoms and nasal eosinophil counts in allergic rhinitis patients [66]. However, the data remain limited. Further studies are needed to fully understand the role of nasal microbiome dysbiosis in allergic rhinitis. Although the correlation between nasal microbiota and asthma is poorly defined, several recent studies have demonstrated that the nasal microbiota plays a significant role in asthma onset, development, and severity [67-69]. One study reported on the nasopharyngeal microbiome during the first year of life and found that the nasopharyngeal microbiome was a determining factor of future asthma development and the severity of its accompanying inflammatory symptoms, with early asymptomatic colonization with Streptococcus in particular being a strong predictor of the future risk of asthma development [65]. Another study reported that the composition and structure of the nasal microbiota of children and adolescents with asthma were significantly different from those of healthy controls and varied among different asthma phenotypic clusters [70]. In asthmatic children. the nasal microbiome was dominated by Moraxella, Staphylococcus, Corynebacterium, Haemophilus, Fusobacterium, Prevotella, and Dolosigranulum [68]. Furthermore, asymptomatic colonization in neonates with the common respiratory tract pathogens H. influenzae, M. catarrhalis, and S. pneumoniae has been shown to confer a higher risk of subsequent lung diseases, including childhood asthma [32,33,71]. One study reported that nasopharyngeal Bordetella pertussis colonizing infections are harmless but that subclinical B. pertussis colonization is an important cause of asthma and allergic sensitization diseases [72]. On the other hand, Bacteroidetes and Proteobacteria, Prevotella buccalis, and Gardnerella vaginalis were abundant in adult asthma patients, and P. buccalis, G. vaginalis, Dialister invisus, and Alkanindiges hongkongensis species were differentially abundant depending on asthma activity [73]. However, future studies are needed of nasal microbiome dysbiosis in asthma patients.
Novel approaches to controlling nasopharyngeal microbiota
Although antibiotics and vaccines can partially reduce or eliminate nasal pathogens, they cannot correct nasal microbiota dysbiosis. Human pathogens, which are also part of the normal microbiota, complicate the eradication paradigm. There have been recent promising developments in the use of probiotics or probiotics with prebiotics as adjuvant treatments for controlling nasal dysbiosis. Several studies have reported that some probiotics such as Lactobacillus acidophilus, Lactobacillus paracasei, and Lactobacillus rhamnosus GG improve symptoms of allergic rhinitis by altering immunologic responses [74-76]. In 2 separate trials, newborns who were administered synbiotics had a significantly lower incidence of respiratory tract infections (especially rhinoviral infections) than those administered placebo [77,78]. Probiotics such as Streptococcus salivarius 24SMBc and Streptococcus oralis 89a may reduce S. aureus as a result of limiting the overgrowth of potential pathogens [79]. The intranasal administration of Bacillus subtilis vaccine in humans to enhance the immunity of human nasal mucosa to respiratory diseases could be attempted in the future (Table 3) [80].
Conclusions
The nasal microbiota varies with age and is shaped by various influencing factors in healthy individuals. A pathological condition of the respiratory tract appears to be associated with a reduction in nasal microbiota biodiversity; a similar feature was also shown in other sites such as the gastrointestinal tract. Nasopharyngeal microbiota dysbiosis is involved in the pathophysiology of many respiratory diseases, including OM, sinusitis, allergic diseases, and lower respiratory tract infections. In addition, recent studies suggested that the profile of nasal microbiota influences immune responses and has the potential to be used as a prognostic tool for diseases in clinical practice. Therefore, instead of previous classical culture approaches and high-cost methods, the use of widely available low-cost methodologies should be considered to enable the practical clinical use of the nasal microbiota profile. For instance, various simple polymerase chain reaction methods, such as broadrange polymerase chain reaction that uses the 16S rRNA to identify taxonomy of bacterial species, are relatively cheap, can only identify a relatively small array of genes, and are useful in identifying disease biomarkers and prognostic factors in clinical care.
Of note, current data in the literature were acquired from cross-sectional cohort studies involving diseased and healthy populations; therefore, longitudinal cohort studies are required. In addition, we should consider the importance of a combined classification model including bacterial microbiome, viral microbiome, and external factors such as antibiotic use and breastfeeding in predicting clinical disease status. Several studies suggested that probiotic or synbiotic interventions may show promising effective roles in the adjunctive treatment of perennial allergic rhinitis and seasonal allergic rhinitis, with improvements occurring through species-specific interactions and immunological modulatory responses (Table 3). However, data are limited and future studies are needed to characterize how these interventions can be applied in clinical practice.





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link PubMed
PubMed Download Citation
Download Citation