Introduction
Influenza has been occurring every year, on a large scale. Thus, it may cause huge socioeconomic loss. In addition, due to the possibility of the appearance of novel viruses every year caused by frequent antigenic variation of virus, it may cause high mortality
1). Accordingly, antigenic variation of the influenza virus has been continuously monitored via a worldwide surveillance program to develop vaccines and to investigate pandemic status
2).
Preventive vaccination is important for influenza prevention. In addition to the preventive vaccination, antiviral agents also play an important role in the treatment of influenza
3). Amantadine and rimantadine, M2 inhibitors, are ineffective in the influenza B virus infection, without the M2 protein, due to a rapid increase of drug resistance and side effects in the central nervous system; their use is limited in pediatric patients
4,
5).
Oseltamivir (Tamiflu), a neuraminidase (NA) inhibitor, has been approved for the prevention and treatment of influenza A and B in pediatric patients, aged one year or higher and in adult patients, and has been used worldwide since 2001
3). However, few studies on oseltamivir have been conducted on Korean pediatric patients. Accordingly, the authors randomly divided the influenza patients during 2007-2008 and 2008-2009 influenza seasons. In to the oseltamivir treatment group and oseltamivir nontreatment group, and investigated the efficacy, short-term and long-term adverse events, and safety of oseltamivir treatment. As a result, the authors showed that oseltamivir treatment had clinical efficacy and safety in pediatric patients by shortening hospitalization duration and reducing lower airway complications without other serious adverse events including neuropsychiatric problems for both 2007-2008 and 2008-2009 influenza seasons
6).
However, since its commercialization, increase in oseltamivir resistance by virus has been reported since 2004
7,
8), and has been reported worldwide since 2007-2008
9). In addition, in Korea, influenza sample surveillance during 2008-2009 influenza season showed that oseltamivir resistance was shown in 99.7% of A/H1N1 virus isolates
10). The occurrence of oseltamivir-resistant virus may decrease chemoprophylaxis and efficacy for the influenza virus. In particular, drug-resistant virus occurs more frequently in pediatric patients than in adult patients, due to limited immunity and long-term virus release in children
8,
11), and the occurrence of drug-resistant virus in children not only affects the individual pediatric patient, but also becomes a very important issue in terms of public health. Accordingly, this study was conducted to investigate the occurrence and resistance mechanism of oseltamivir-resistant virus between the oseltamivir treatment and non-treatment groups, during 2007-2008 and 2008-2009 influenza seasons.
Materials and methods
1. Subjects, oral administration of oseltamivir, and disease history review
The authors previously reported a clinical study on the efficacy, short-term and long-term side effects, and safety of oseltamivir in pediatric patients infected with influenza virus
6). The summary of the clinical study was as follows. Of the pediatric patients with respiratory symptoms who visited the outpatient and emergency room in the department of pediatrics at Busan St. Mary's Medical Center, during the two influenza seasons (9 months), January 2008 to June 2008 (first study period) and November 2008 to January 2009 (second study period) those with influenza-like symptoms were selected. As for the influenza-like symptom, it was defined as a status of 37.8℃ or higher fever as a main symptom with one or more systemic symptoms (fatigue, headache, muscle pain, chill) and respiratory symptoms (cough, pharyngitis, nasal discharge, nasal congestion) by referring to the definition of the National Institute of Health
12). But, children of less than 2 years old age were enrolled into the study if they had an temperature ≥37.8℃ and at least one respiratory symptoms (cough or coryza).
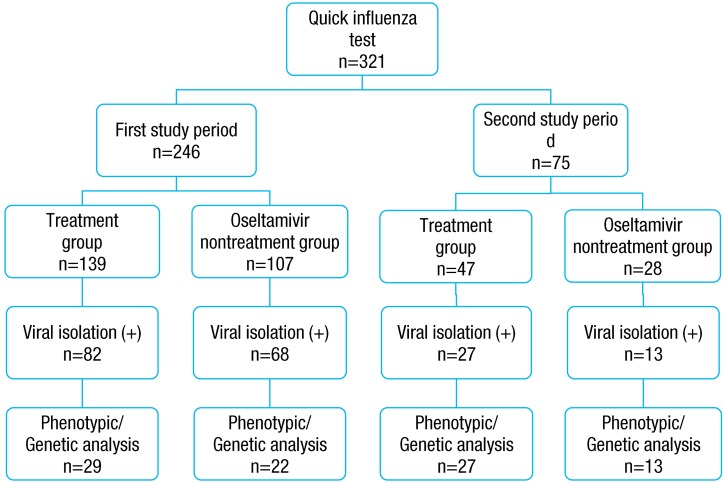
Nasal discharge was collected from the patients, and then quick influenza test (SD Bioline Influenza Antigen Test kit; Quidel Co., San Diego, CA, USA), a rapid antigen test (RAT), was conducted to diagnose the influenza. The same tests were performed during two periods. The RAT positive in all patients, a total of 537 pediatric patients (first study period 318, second study period 219) who were diagnosed as positive to influenza and hospitalized. All parents of influenza patients provided written informed consent.
The study was prospectively conducted after receiving the informed consent forms from the participating patients. Among parents of 537 patients, 321 parents' consent and their child participated in the study and assigned to the oseltamivir treatment group (A group) and oseltamivir nontreatment group (B group) based on the assigning to the each attending doctor. Study exclusion criteria included: children with respiratory syncytial virus infection (RAT); cyanotic heart disease; children with immunosuppressed (drugs, transplant recipient, malignancy or human immunodeficiency virus infection); history of acetaminophen allergy documented. Throat swab or nasopharyngeal swab was collected from the RAT positive in all patients, upon admission and 48 to 72 hours after admission, tests were performed two times, and transferred to Busan Institute of Health and Environment for virus culture. Immediately after virus culture, the A group received oseltamivir, antiviral agent, and twice a day for 5 days. Meanwhile, the B group received symptom controlling agent when they had influenza-like symptoms. The treatment dose was 30 mg for body weight of 15 kg or less, 45 mg for body weight of 16 to 23 kg, and 60 mg for 24 to 40 kg, and 75 mg for body weight of 40 kg or more in patients aged 1 to 12 years. The drug was administered twice a day for 5 days. Meanwhile, the drug was administered at a dose of 75 mg, twice a day for 5 days, in the pediatric patients aged 13 years or higher.
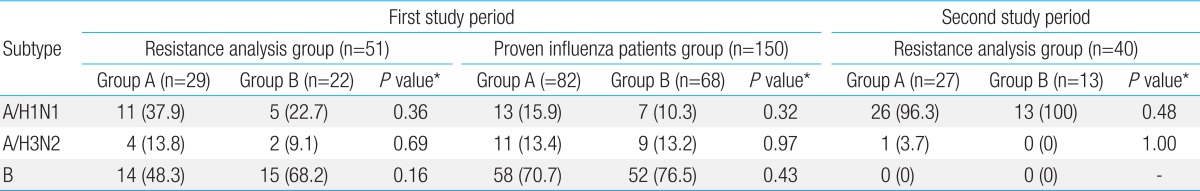
Of the 321 pediatric patients, who were hospitalized due to influenza, viruses were identified by culture in 190 patients (first study period 150 patients, second study period 40 patients). Drug resistance test was conducted on 91 patients (first study period 51 patients, second study period 40 patients) among the 190 culture proven patients. However, in the first study period, drug resistance test was conducted by 20% random sampling in the Centers for Disease Control because of limitation of laboratory environment. The pediatric patient's sex, age, preventive vaccination taking and clinical data including fever and admission duration were reviewed, using their medical records. The study has been approved by the Institutional Review Board of Busan St. Mary's Medical Center before the study initiation.
2. Methods of virus detection
1) Culturing and RNA analysis of influenza virus
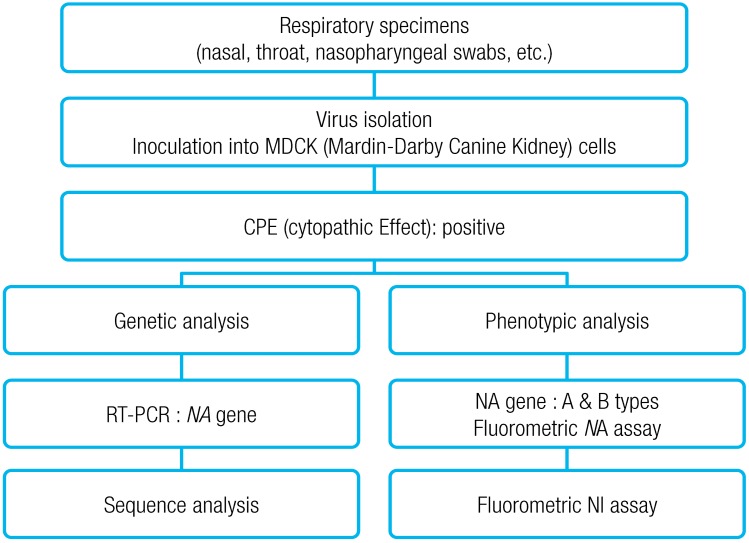
Throat and nasopharyngeal swabs were collected using a sterilized cotton stick from the pediatric patients who were hospitalized after being diagnosed as positive to influenza as a result of a RAT, and stored in a refrigerator. They were put into a viral transport medium (VTM; Difco Laboratories, Detroit, Ml, USA), and then transferred to Busan Institute of Health and Environment, while maintaining a 4℃, followed by virus culture. Mardin-Darby Canine Kidney (MDCK) cell line, provided by the department of influenza virus of the Center for Disease Control and Prevention (CDC), was cultured on minimum essential medium (MEM; Gibco, Carlsbad, CA, USA), containing penicillin (0.05 U/mL), streptomycin (0.05 µg/mL), and 10% fetal bovin serum (FBS; Gibco USA) in a 5% CO2 incubator at 37℃. For specimen pretreatment, penicillin (5 U/mL), streptomycin (5 µg/mL), and nystatin (1,000 units/mL) were added to the VTM, and were placed at 4℃ for one hour, while shaking at an interval of 15 minutes, followed by centrifugation (2,000 rpm, 20 minutes, 4℃). The supernatant obtained from the centrifugation was used as a specimen for virus inoculation. For virus isolation, 200 µL pretreated specimen and MDCK inoculation culture solution 500 µL were multiply inoculated on 3 wells of the MDCK cell line that was monolayer-cultured in a 24 well culture vessel, followed by culturing in a CO2 incubator at 5% CO2, 35℃ for 7 to 10 days. The cells were observed on a daily basis, under an inverted microscope. If 80% or more cytopathic effect (CPE) was observed, the cells were collected and then centrifuged (10,000 rpm, 10 minutes, 4℃) to separate the supernatant and cell sediments, and were stored at -70℃ before use. If necessary, they were sequentially cultured 2-3 times to increase the titer, and the culture solution and infected cells were obtained for virus isolation and identification. Virus identification was performed via hemagglutination inhibition test and reverse transcriptionpolymerase chain reaction (RT-PCR). For RT-PCR, RNA was isolated from the virus culture solution with CPE, using a ZR viral RNA kit (Zymo Research Co., Irvine, CA, USA), followed by RT-PCR to analyze the virus subtype (A/H1N1, A/H3N2, and B).
2) Antiviral agent resistance test
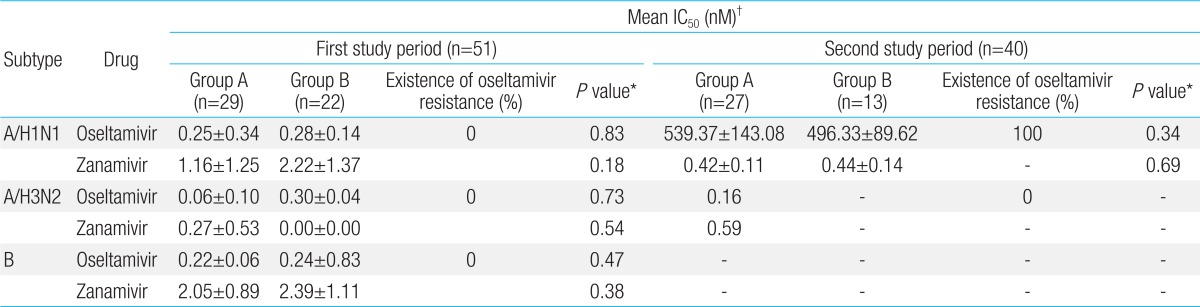
Influenza antiviral agent drug resistance test was conducted via genotypic and phenotypic analyses (
Fig. 1). For the phenotypic analysis, NA inhibitors (oseltamivir, zanamivir) were used, according to fluorometric NA inhibition assay (World Health Organization [WHO]-025), which is the standard assay provided by the WHO Collaborating Centre for Reference and Research on Influenza. After NA inhibitor was prepared to have a concentration of 0-30,000 nM, 50 µL of the NA inhibitor was put into a 96 well black plate. Influenza virus of 50 µL was added to each well, and 1× AB 50 µL was added to the last well, as the control, followed by a reaction at RT for 45 minutes. 50 µL methylumbelliferone N-acetylneuraminic acid was added to each well, and then reacted at 37℃ for 1 hour, the reaction was halted by adding 100 µL stop solution, and absorbance was measured at 360 nm and 448 nm. Amount of NAI required to inhibit 50% of viral NA activity, 50% inhibitory concentration (IC
50 [nM]) values for each virus was obtained, using a Prism 4.0 program. According to the criteria presented in the Australian Influenza Reference Laboratory the virus was adjudged to be a sensitive strain in the case of IC
50 value of 0.001-25 nM, and resistant strain in the case of IC
50 value of 43-8,020 nM.
For the genotypic analysis, mutations of amino acid regions related to drug resistance (E119V, R152K, H274Y, R292K, and N294S) were examined, via sequence analysis of NA gene. RT-PCR was conducted using primers specific for NA gene of influenza viruses isolated during the second study period and RT-PCR premix (AccuPower RT/PCR PreMix, Bioneer, Daejeon, Korea) according to Jung's method (2008)
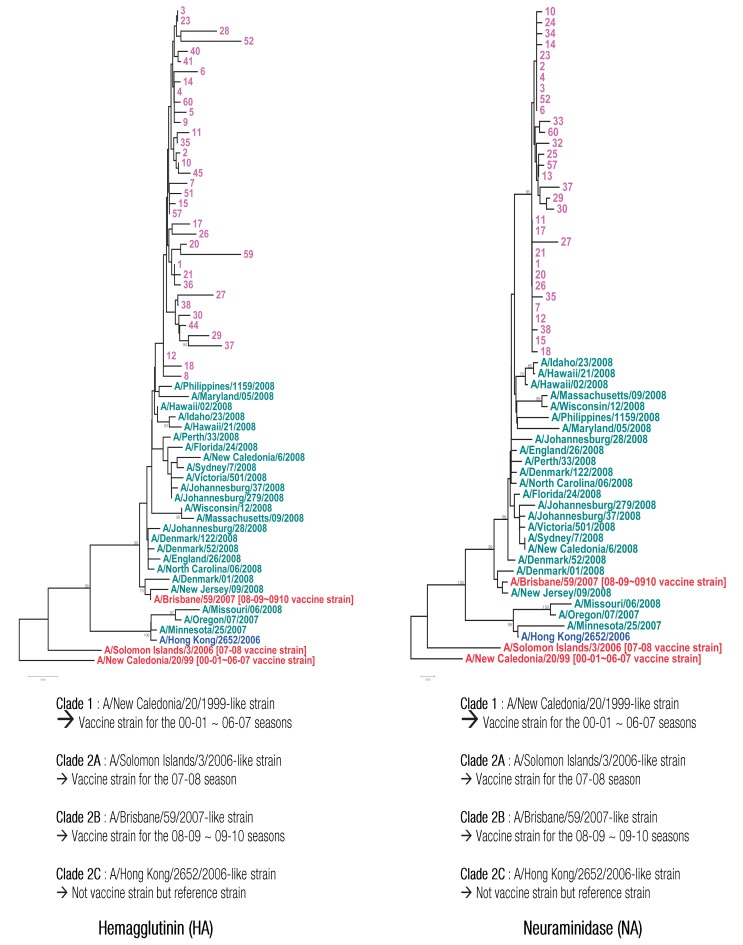
13). That is, strand RNA and reverse primer mixture were reacted at 70℃ for 5 minutes, and then immediately transferred on the ice, followed by a reaction for 5 minutes. Then, forward primer was added and reacted at 42℃ for 1 hour to prepare cDNA. Subsequently, PCR was conducted under the conditions of 94℃ for 1 minute, 61℃ for 1 minute, 72℃ for 1 minute, and 35 cycles. The PCR product underwent electrophoresis at 1.5% agarose gel (0.5×tris-acetate-ethylenediaminetetraacetic acid) to identify NA gene-specific PCR product. The PCR product was purified using PCR purification (Bioneer) and gel extraction kit (Qiagen, Valencia, CA, USA), followed by nucleotide analysis using ChromasPRO, EditSeq and MegAlign (DNASTAR, Madison, WI, USA). PAUP ver. 4.0 was used for genetic distance and phylogenetic tree. A comparative analysis was conducted using the finally identified nucleotide sequences among the WHO recommended vaccines, foreign isolates, and domestic isolates.
3. Statistical analysis
Statistical analysis was conducted using the SPSS ver. 12.0 (SPSS Inc., Chicago, IL, USA). Continuous values were expressed as the mean and standard deviation. Chi-square test was conducted for analyzing qualitative factors, and independent t-test, Mann-Whitney, and Kolmogorov-Smirnov tests were conducted for the analysis of quantitative factors, depending on the presence of normal distribution in the data. If P value was <0.05, it was considered statistically significant.
Discussion
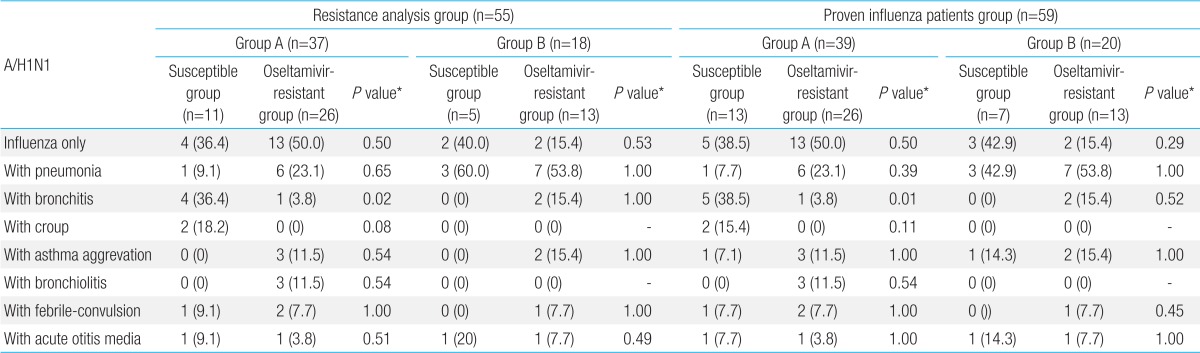
In this study, influenza A/H1N1 isolated from both treatment group and oseltamivir nontreatment group during the first study period (2007-2008 season) and second study period (2008-2009 season) had no zanamivir resistance. However, phenotypic analysis showed that oseltamivir IC50 ranged within high-level drug resistance, H275Y of NA gene and partial variation of HA gene does not affect antigenicity to HA vaccine, even though oseltamivir treatment group had shorter admisson duration and fewer lower respiratory tract complications compared to the oseltamivir nontreatment group. In addition, there was no significant difference of clinical manifestations between oseltamivir susceptible and resistant periods.
As for the clinical characteristics of treatment group and oseltamivir non-treatment group during the first study period or second study period, no significant difference in age, gender, presence of preventive vaccination, and previous history of influenza infection was found between the two groups. The result of influenza virus isolation showed that influenza virus B, A/H1N1, A/H3N2 in that order were isolated in treatment group and oseltamivir nontreatment group during 2007-2008 influenza season, and that influenza A/H1N1 was mainly isolated in the both groups during 2008-2009 influenza season. This result was consistent with the subtype distribution of 2007-2008 and 2008-2009 influenza sentinel surveillance reported by the center for disease control
10).
New influenza occurs each year via antigenic shift and antigenic drift due to genetic changes of
HA and
NA genes, surface antigens. Antigenic shift mainly occurs by influenza A, and is classified into H1-H16 according to HA mutation, and into N1-N9 types according to NA mutation
14). HA has been known to be a main causative factor of antigenic mutation and involved in the binding of virus to cells, and NA has been known to be a surface antigen of influenza virus and play an important role in the secretion of synthesized viruses from cells, thus, they have drawn attention as targets of antiviral agents
3). Oseltamivir, one of NA inhibitors, is an oral agent that is used worldwide for the prevention and treatment of influenza. However, since a rapid increase of oseltamivir-resistant influenza virus A/H1N1 was reported in Norway in January 2008, oseltamivir-resistant strains have been spreading worldwide including Europe, Oceania, Southeast Asia, and Africa
9,
15,
16). In this study, no oseltamivir resistance was shown in any influenza subtype during the 2007-2008 season in but 100% oseltamivir resistance was shown in all the influenza A/H1N1 strains during the 2008-2009 season in accordance to domestic data
10). The results of genotype analysis revealed H275Y (N1 numbering) mutation, which was consistent with those of previous studies
9,
16). In addition, the result of phenotype analysis showed that zanamivir IC
50 ranged within sensitivity, but that oseltamivir IC
50 ranged beyond the maximum concentration, showing oseltamivir resistance. This was also consistent with those of the previous studies
17).
Oseltamivir-resistant virus before 2007-2008 season was mainly reported in pediatric patients who received oseltamivir
8), and human to human transmission has been rarely reported
18). However, in the cases of pediatric patients infected with oseltamivir-resistant virus isolated in US during 2007-2008 season and in Italy during 2007-2008 and 2008-2009 seasons, they had no previous history of oseltamivir administration
19,
20). In this study, although the previous history of oseltamivir administration was not checked in all the pediatric patients, most of pediatric patients during 2008-2009 influenza seasons had no previous history of oseltamivir administration. Thus, the occurrence of oseltamivir-resistant virus during 2008-2009 seasons is unlikely to be relevant to oseltamivir administration.
In Japan, the rate of oseltamivir-resistant influenza A/H1N1 was shown to be 2.5% during 2007-2008 season, but 100% during 2008-2009 season
21). Matsuzaki et al.
21) reported that when the genetic analysis of phylogenic tree was conducted, oseltamivir-resistant A/H1N1 that was prevalent in Japan during 2007-2008 season was different from oseltamivir-resistant A/H1N1 that was prevalent in Europe and North America during 2007-2008 season, but oseltamivir-resistant A/H1N1 that was prevalent in Japan during 2008-2009 season showed high homology with oseltamivir-resistant A/H1N1 that was prevalent in Europe and North America during 2007-2008 influenza season. In this study, oseltamivir-resistant influenza A/H1N1 type during 2008-2009 seasons had H275Y mutation of NA gene and D354G mutation, which was genetically close to oseltamivir-resistant A/H1N1 that was prevalent in Europe and North America during 2007-2008 season
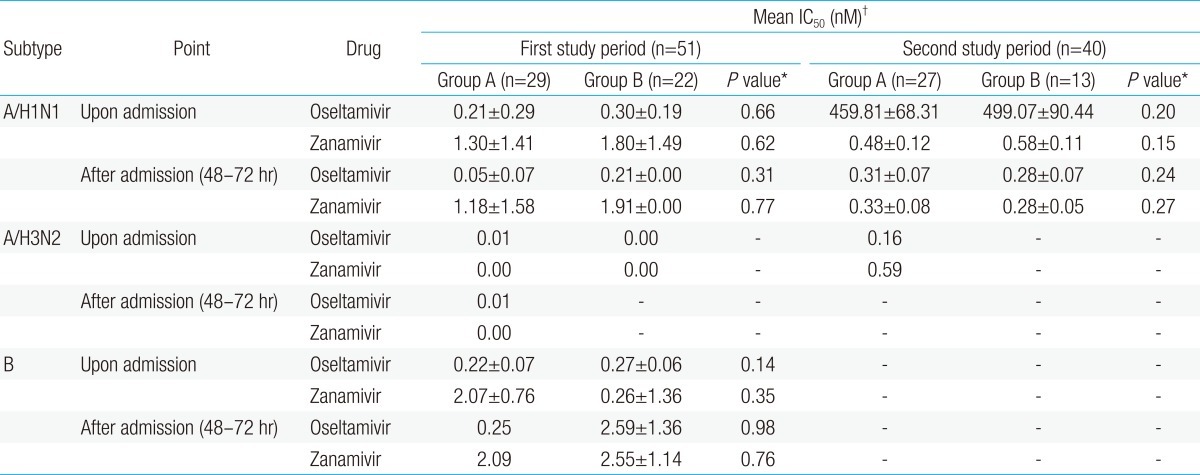
9). In addition, the analysis of virus culture isolated upon admission and 48 to 72 hours after admission showed that the occurrence rate of oseltamivir-resistant influenza was zero both in the treatment group and in the oseltamivir nontreatment group during 2007-2008 season. The occurrence of oseltamivir-resistant influenza A/H1Nl during second study period was unlikely to have occurred due to oseltamivir administration during first study period. It is likely that oseltamivir-resistant influenza A/H1N1 that was prevalent in Europe and North America during 2007-2008 influenza seasons have been imported to Japan or Korea like the results of other studies
21). Esposito et al.
20) studied pediatric patients who were infected with influenza during 2007-2008 and 2008-2009 seasons, and reported that the H275Y mutation rate of influenza A/H1N1 was 1.6% during 2007-2008 influenza season, and 100% during 2008-2009 influenza season, and they also reported that no difference in the patient's characteristics and hospitalization duration was found between pediatric patients infected with oseltamivir-resistant influenza A/H1N1 and those infected with wild-type seasonal A/H1N1 in compatible with this study . The authors' previous study also reported that no significant difference in the clinical manifestations of the patients was found between 2007-2008 influenza season when no oseltamivir-resistant influenza was shown and 2008-2009 influenza season when oseltamivir-resistant influenza A/H1N1 was 100% shown
6). However, Dharan et al.
19) reported that despite the similar clinical manifestations of patients infected with oseltamivir-resistant influenza A/H1N1 and with oseltamivir-susceptible influenza A/H1N1, four patients of the 99 patients infected with oseltamivir-resistant influenza A/H1N1 died, and that oseltamivir-resistant influenza A/H1N1 had serious clinical manifestations, such as death. They also reported that one of the four dead patients was a previously healthy female patient aged 4 years, and that severe clinical manifestation with oseltamivir-resistant influenza infection could be found even in healthy children.
The authors' previous study
6) reported that compared to the oseltamivir nontreatment group, shorter admission duration and fewer lower respiratory tract complications occurred in the treatment group during the 2007-2008 season with no occurrence of oseltamivir-resistant influenza A/H1N1, and the 2008-2009 season with oseltamivir-resistant influenza A/H1N1 in accordance to this study.
Mitamura et al.
22) reported that oseltamivir treatment was initiated within 48 hours of the onset of fever, 44% of the patients became afebrile (<37.5℃) within one day, and 86% of them recovered within two days and the average duration of fever after the initiation of oseltamivir treatment was 1.7 days in contrast to this study. We suggested that frequency of duration of time between fever onset and admission point is less than 48 hours was lesser in oseltamivir treatment group compared than oseltamivir nontreatment group in this study and effectiveness of oseltamivir treatment against influenza virus infection in children was attenuated and underestimated.
As no difference in the fever duration was found in the treatment group, between the first (oseltamivir-susceptible period) and second study periods (oseltamivir-resistant period) after oseltamivir administration in both previous study
6) and this study, it seemed that no significant difference in the efficacy of oseltamivir between the first study period and second study period where oseltamivir-resistant virus appeared. However, Matsuzaki et al.
21) reported that oseltamivir efficacy could be reduced more in the oseltamivir-resistant strains as the fever persistency after oseltamivir administration was longer in the patients infected with oseltamivir-resistant influenza A/H1N1 than in the patients infected with oseltamivir-susceptible influenza A/H1N1. Tamura et al.
23) reported that virus shedding time was shorter in pediatric patients who underwent zanamivir treatment than in pediatric patients who underwent oseltamivir treatment, in a study conducted on 144 pediatric patients during 2005-2006, 2006-2007, 2007-2008, and 2008-2009 influenza seasons, and that no zanamivir-resistant strain was found in the patients who underwent zanamivir treatment, and they concluded that zanamivir was better than oseltamivir in the treatment of pediatric influenza. Currently, CDC and American Academy of Pediatrics define oseltamivr and zanamivir, which are NA inhibitors, as the therapeutic agents for influenza A and B, including 2009-2010 pandemic H1N1 strain in the cases of influenza-infected high-risk group, who might have influenza complications, including pediatric patients aged less than 2 years, child hospitalized with presumed influenza, and children with confirmed or suspected influenza who have severe, complicated, or progressive illness
24,
25). Thus, a further study on efficient treatment strategies on pediatric influenza is required.
In addition, unlike the 2007-2008 season, the influenza A/H1N1 isolated during 2008-2009 season had high oseltamivir IC
50 in the phenotypic analysis, and had H275Y (N1 numbering) mutation in the genotypic analysis, showing that NI-resistant virus was prevalent. As they were genetically close to oseltamivir-resistant virus, reported in Europe and North America during 2007-2008 seasons, oseltamivir-resistant influenza A/H1N1 virus seems to have been rapidly made its way to Korea during the two influenza seasons. Although the exact mechanism of the importing of oseltamivir-resistant influenza to Korea is difficult to elucidate in this study, it is likely that due to the mutation of
NA gene and mutation of
HA gene, immunogenicity to oseltamivir-resistant virus as well as antibody formation have decreased
26).
Although there were some amino acid variations (158, 202, 206) in
HA gene of oseltamivir-resistant influenza A H1N1 virus identified during 2008-2009 study period, it was confirmed that there was no change of antigencity of strain due to these amino acid variations based on data of hemagglutination inhibition of domestic and foreign strains
27). Specially, because that A/Brisbane/59/2007 was used as vaccine strain during 3 years before outbreak of H1N1 2008/2009, we assumed that there was no considerable effect on antigenicity of vaccine strain based on genetic and serological analysis during this period. In addition, as the analysis of
HA gene of national NI-resistant viruses showed that all of them were similar to vaccine of the corresponding year; efficient prevention can be performed via vaccination.
Considering that the frequency of drug-resistant influenza virus is higher in children than in adults, and that the occurrence of these drug-resistant influenza viruses affects not only the individual health of children, but also public health, this study is meaningful in that it was a first study prospectively conducted on Korean children, despite a disadvantage of a single institution study. In addition, the results of this study could be useful for establishing more detailed monitoring on drug-resistant influenza virus that occurs continuously, and for establishing treatment strategies for treating pediatric patients infected with annual prevalent influenza.
In conclusion, based on the our knowledge, this study was the first study to investigate Korean pediatric patients infected influenza virus (two groups based on the oseltamivir treament), their oseltamivir resistance, NA H275Y protein variation, partial variation of HA in the influenza A/H1N1 virus isolated from both oseltamivir treatment and non-treatment groups, during the progression from the first study period to the second study period even though oseltamivir treatment group had shorter admission duration and fewer lower respiratory tract complications compared to the oseltamivir nontreatment group. In addition, clinical manifestation was no significant difference between oseltamivir-susceptible and resistant periods.
Therefore, the establishment of a guideline on the efficient treatment of influenza, using oseltamivir that is commonly used for treating influenza in Korean pediatric patients and the establishment of treatment strategy using a new therapeutic agent are required.






 PDF Links
PDF Links PubReader
PubReader PubMed
PubMed Download Citation
Download Citation